Multiple Sequence Alignments
We performed two multiple sequence alignments, one comparing the isoforms within humans and one comparing Human isoform 1 with other species.
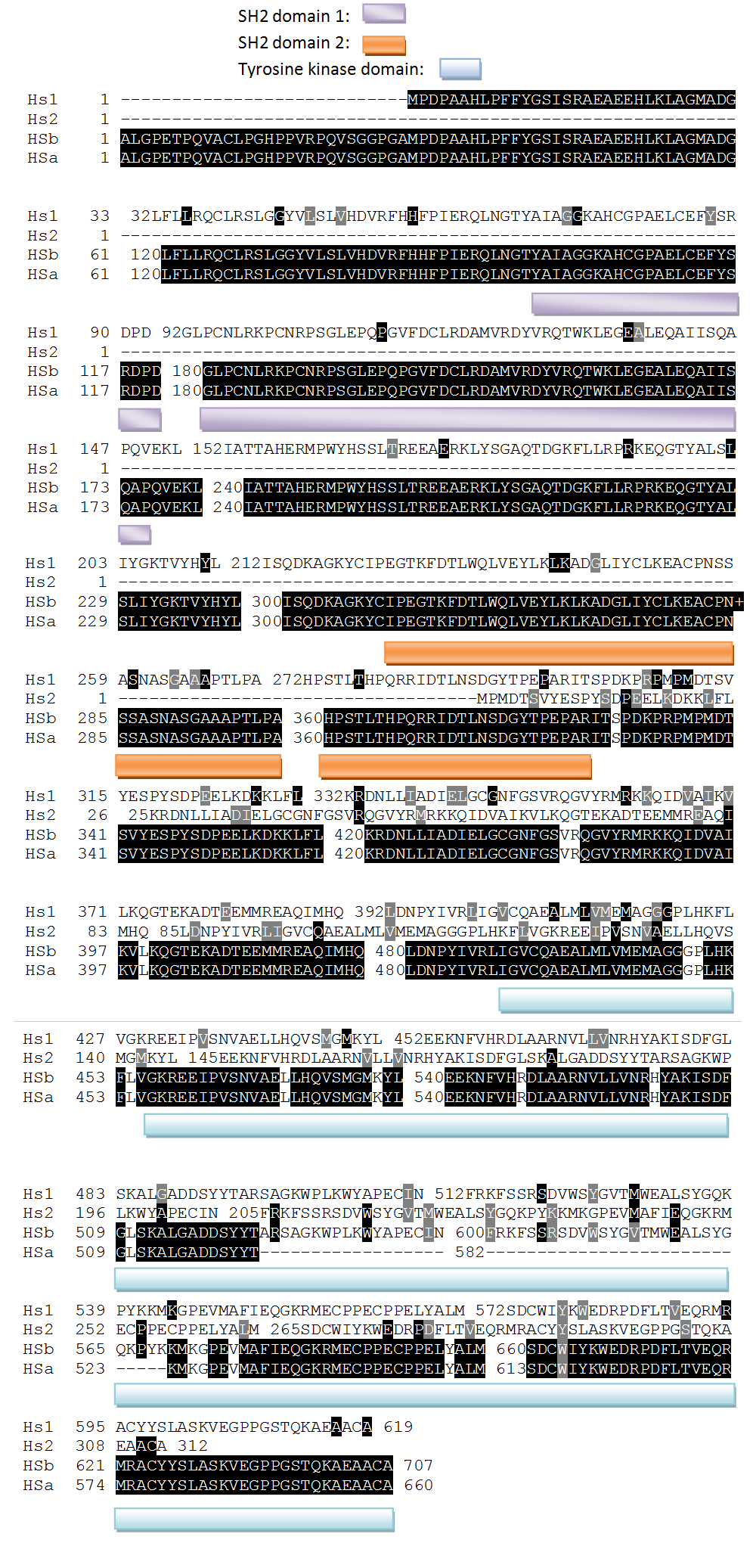
Human Isoform Multiple Sequence Alignment
All the four human isoforms have highly conserved SH2 domains, HS2 has no tyrosine kinase domain and Hsa and Hsb have exactly the same SH2 domains and similar tyrosine kinase domains.
| SH2 Domains | Tyrosine Kinase Domains | |
|---|---|---|
| Isoform 1 (Hs1) | 10-87 & 163-238 | 342-591 |
| Isoform 2 (Hs2) | 35-285 | - |
| CRA a (Hsa) | 98-175 & 251-326 | 430-584 |
| CRA b (Hsb) | 98-175 & 251-326 | 430-679 |
Human Isoform 1 vs Other Species Alignment
This is the multiple sequence alignment of Hs1 ZAP-70 against ZAP-70 homologues in other mammals, amphibians and fish. Compared to Hs1 it can be seen that the ZAP-70 homologues have similar highly conserved SH2 domains and tyrosine kinase domains. Apart from the domains, there are conserved amino acid sequences in all the proteins. The active sites, located in the catalytic domain (tyrosine kinase domain) of the homologues, vary across species and can be seen in the BLAST searches.
Canine ZAP-70 is mostly composed of the tyrosine kinase domain, with no SH2 domains and the alignment does not reveal any homologous regions with any of the tyrosine kinase domains except for a tyrosine residue present at position 59, and in a similar position in the other species. The high degree of homology across the various species indicates the importance of ZAP-70 and its homologues in their respective roles. For more information please refer to the next section.
| SH2 Domains | Tyrosine Kinase Domains | |
|---|---|---|
| ZAP70 Isoform 1 [Homo Sapiens] (Hs1) | 10-87 & 163-238 | 342-591 |
| ZAP70 [Mus musculus] (MM) | 10-87 & 163-238 | 341-590 |
| ZAP70 [Bos Taurus] (BT) | 10-87 & 163-238 | 340-590 |
| ZAP70 [Canis lupus familiaris] (CF) | Not applicable here as canine ZAP-70 only has a tyrosine kinase domain. | 3-60 |
| ZAP70 [Xenopus laevis] (XL) | 10-87 & 163-238 | 331-578 |
| ZAP70 [Danio rerio] (DR) | 10-87 & 163-238 | 330-579 |
