Resources
Software and tools developed by the lab
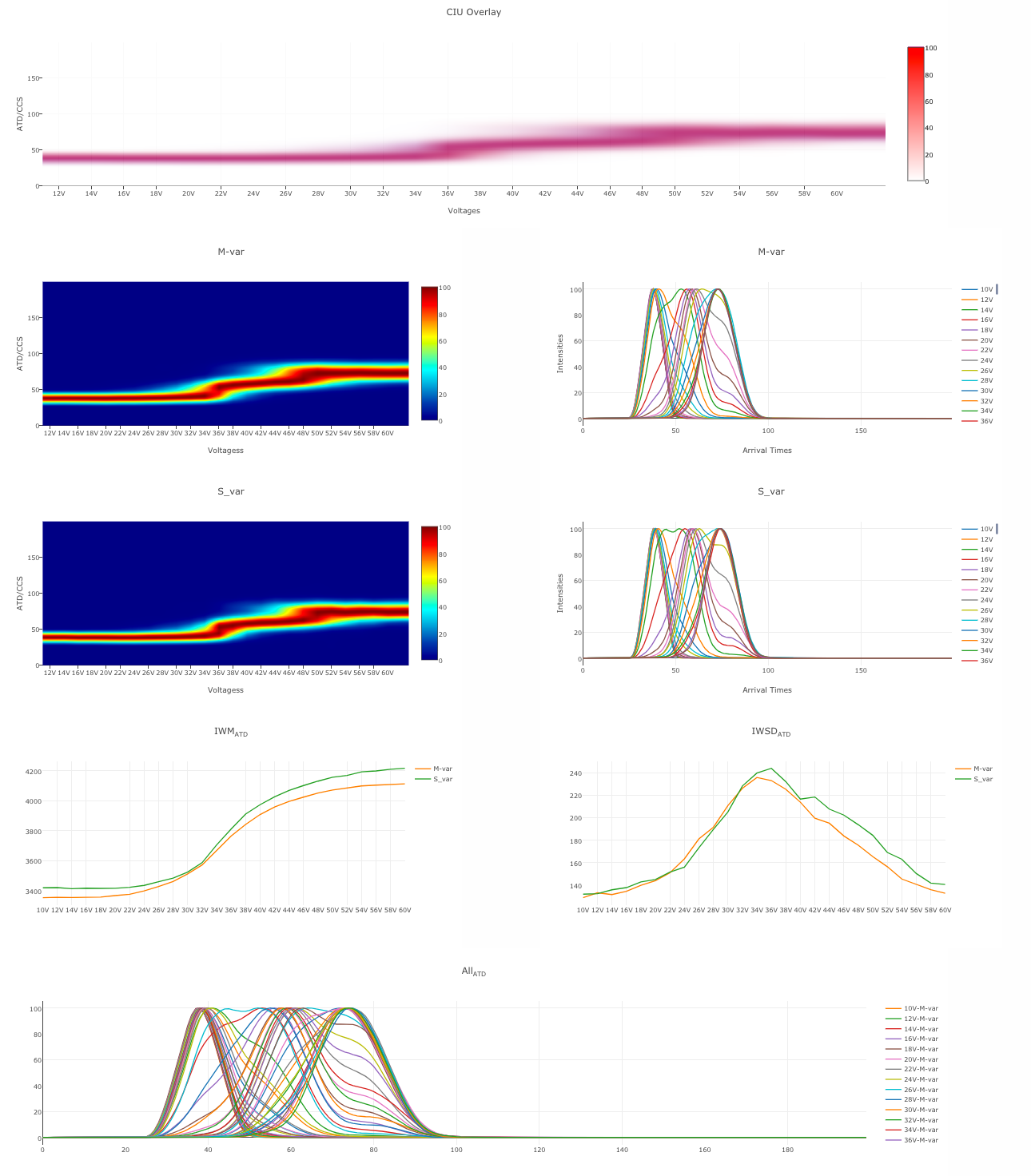
Benthesikyme
A web application that enables the processing of collision induced unfolding (CIU) ion mobility data. The program automatically creates CIU fingerprints and summary plots that capture the increase in collision cross section and the increase in conformational flexibility of proteins during unfolding.
The input data is a tab delimited file with the format as shown in the table below, where the first column contains the name of the experiment followed by the ATD (or CCS) values and each subsequent row representing one ATD for each voltage
The program is written in JavaScript and runs in the web browser you use to access the site. No transmission of your data takes place to our servers, it remains in your own computer. This program is freely distributed. Use at your own risk.
Sivalingam, Ganesh N., Cryar, Adam, Williams, Mark A., Gooptu, Bibek, Thalassinos, K. International Journal of Mass Spectrometry (2018) 426, 29-37
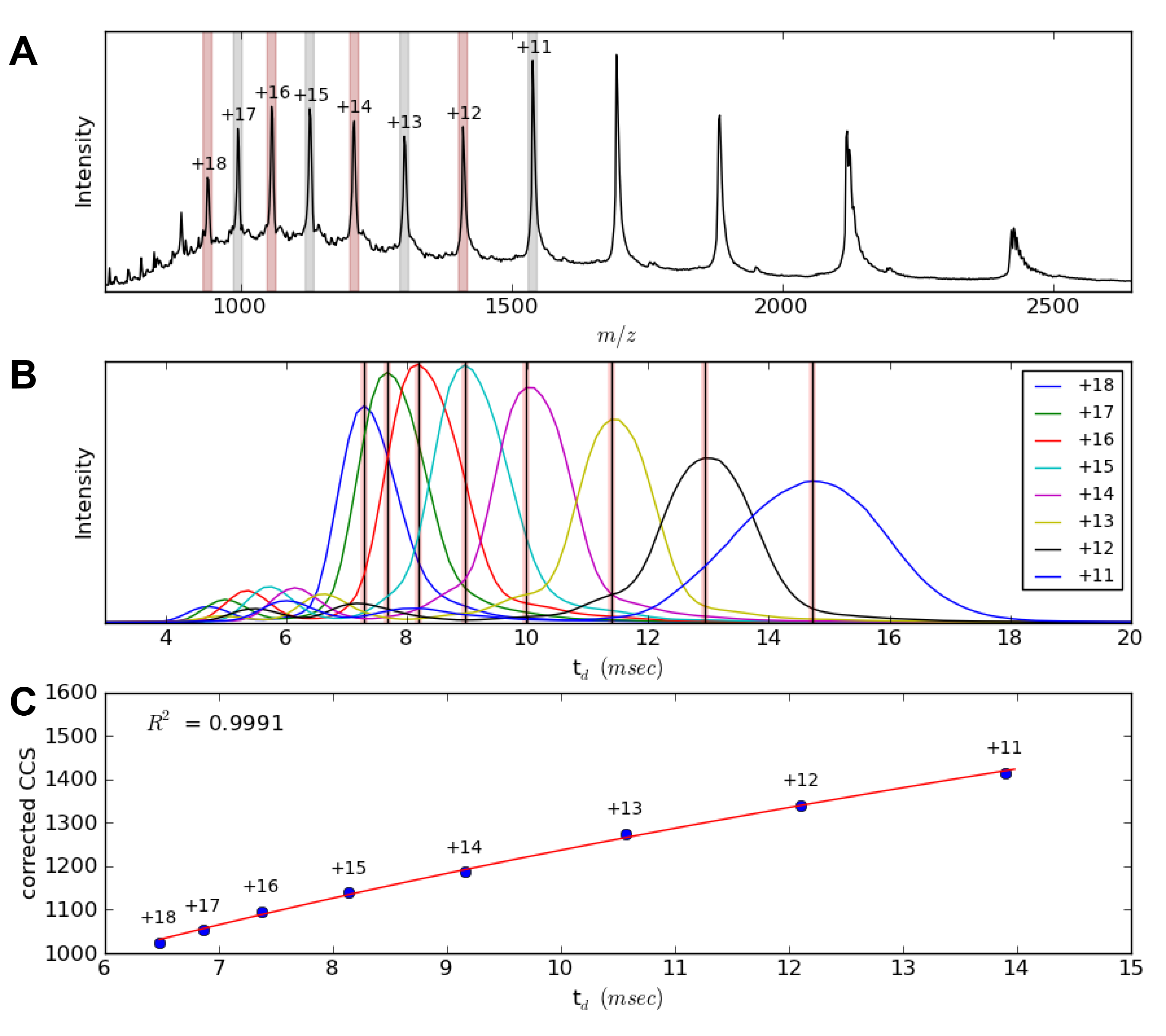
Amphitrite
Amphitrite is a software package that enables the processing of T-Wave ion mobility data. The program can deconvolute components in a mass spectrum and uses this information to extract corresponding arrival time distributions (ATDs) with minimal user intervention. It can also be used to automatically create a collision cross section (CCS) calibration and apply this to subsequent files of interest.
Sivalingam, G.N., Yan, J., Sahota, H., Thalassinos, K. (2013) Amphitrite: a program for processing travelling wave ion mobility data. International Journal of Mass Spectrometry. 345, 54–62.
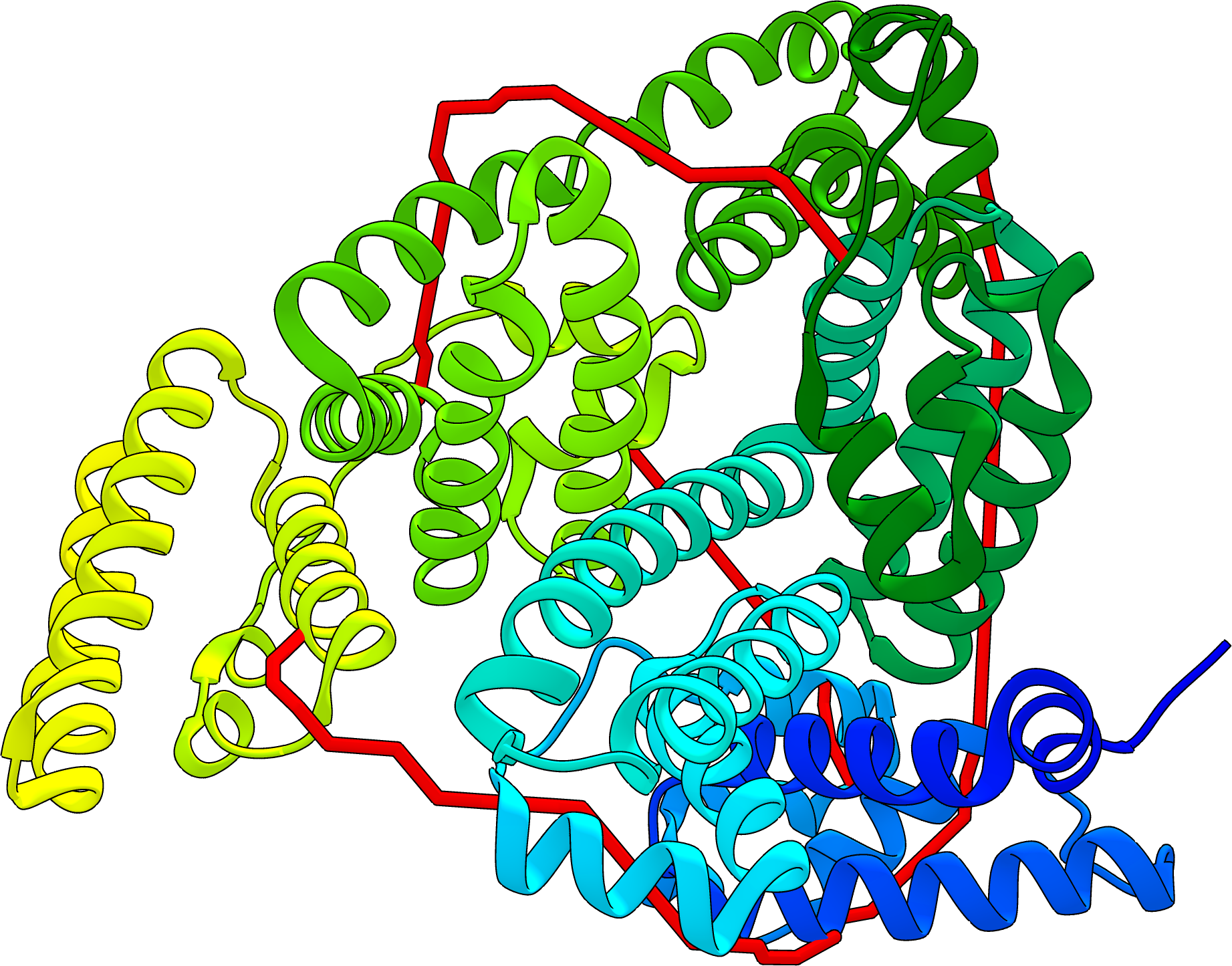
JWalk
Jwalk calculates the distance between crosslinked residues using the two most commonly calculated used metrics: Euclidean Distance or Solvent Accessible Surface Distance (SASD).The Euclidean distance is the direct distance between the two crosslinked residues, however this ignores the fact that a crosslinker cannot travel through protein mass! The more theoretically correct calculation is the SASD. The SASD is defined as the shortest path between two points across the surface of the protein. This metric is computed using a breadth-first search algorithm, which finds the shortest distance between two nodes on a graph.
The Importance of Non-accessible Crosslinks and Solvent Accessible Surface Distance in Modeling Proteins with Restraints From Crosslinking Mass Spectrometry.
J Bullock, J Schwab, K Thalassinos, M Topf. Mol Cell Proteomics. 15, 2491–2500, 2016
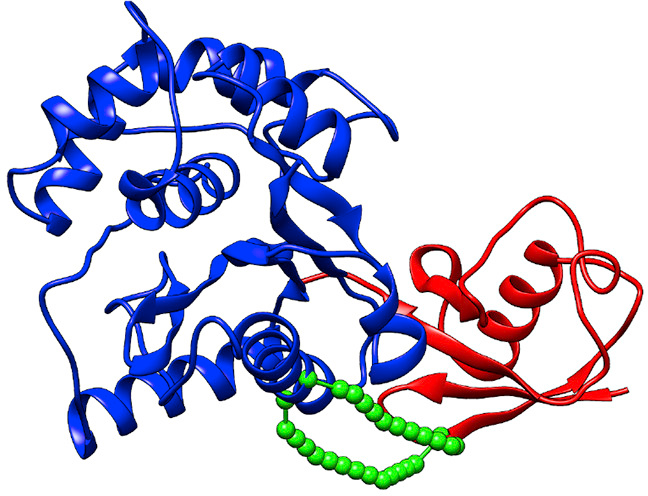
MNXL
Crosslinks contain three different scoring properties: The Expected SASD (Solvent Accessible Surface Area) between two crosslinked residues, The Number of violations of the crosslinker maximum bound, The Number of Non-accessible crosslinks. By considering all three aspects, and utilising the SASD calculated by Jwalk, MNXL (for protein monomers) or cMNXL (for protein complexes) scores and validates models using crosslink restraints. MNXL outperforms more commonly used crosslink validation scores, such as Number of Violations or Sum of Violation Distances. MNXL and cMNXL are only applicable to lysine crosslinkers that are 11.4 Å long (e.g. BS3 or DSS), because the Number of Violations and Expected SASD are dependent on the crosslinker length.
The Importance of Non-accessible Crosslinks and Solvent Accessible Surface Distance in Modeling Proteins with Restraints From Crosslinking Mass Spectrometry.
J Bullock, J Schwab, K Thalassinos, M Topf. Mol Cell Proteomics. 15, 2491–2500, 2016
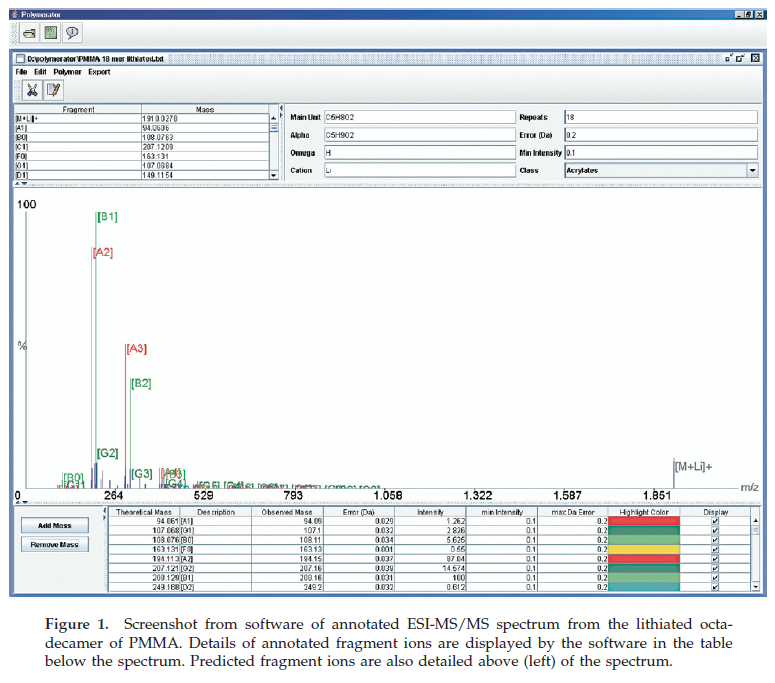
Polymerator
Polymerator is a program that aids the interpretation of tandem mass spectrometry (MS/MS) data from synthetic polymers. The software is particularly focused toward aiding the end-group determination of these materials by significantly speeding up the interpretation process. This allows information on the initiator and/or chain transfer agents, used to generate the polymer, and the mechanism of termination to be inferred from the data much more rapidly. The software allows the validity of hypothesized structures to be rapidly tested by automatically annotating the data file using previously proposed fragmentation rules for synthetic polymers.
This program is freely distributed. Use at your own risk.
Requirements: Java Runtime Environment v1.4.2 or later.
Once downloaded extract the zip file and then double click on the Polymerator-v1.2.jar file in order to start the program.
Thalassinos, K., Jackson, A. T., Williams, J. P., Hilton, G. R., Slade, S. E. and Scrivens, J. H. (2007). Novel software for the assignment of peaks from tandem mass spectrometry spectra of synthetic polymers. Journal of the American Society for Mass Spectrometry 18: 1324-1331.

Synapt Calibration
While the physical principles behind drift cell ion mobility mass spectrometry (IM-MS) are well understood [1] and can be used to obtain a collision cross-section for each ion studied, the same is not true for the T-Wave based device. Methods to create a cross-sectional calibration for the Synapt have been previously published [2-5]. The approach our group has developed is a modified version of Wildgoose et al. [2]. An excel spreadsheet used to simplify the creation and use of a cross-sectional calibration is made available here. A description of our approach is presented in Thalassinos et al. [6].
1) Mason, E. A.; McDaniel, E. W. Transport Properties of Ions in Gases; Wiley: New York, 1988.
2) Wildgoose JL; Giles K; Pringle S; Koeniger S; Valentine S; Bateman RH; Clemmer D. 2006 53rd ASMS Conference on Mass Spectrometry & Allied Topics Seattle.
3) Scrivens, J. H.; Thalassinos, K.; Hilton, G. R.; Slade, S. E.; Pinherio, T. J. T.; Bateman, R. H.; Grabenauer, M.; Bowers, M. T. 2007. 54th ASMS Conference on Mass Spectrometry & Allied Topics.
4) Ruotolo, B. T.; Benesch, J. L.; Sandercock, A. M.; Hyung, S. J.; Robinson, C. V. Nat. Protoc. 2008, 3, 1139-1152.
5) Williams, J. P.; Scrivens, J. H. Rapid Commun. Mass Spectrom. 2008, 22, 187-196.
6) Thalassinos, K., Grabenauer, M., Slade, S. E., Hilton, G. R., Bowers, M. T. and Scrivens, J. H. (2009) Characterisation of phosphorylated peptides using travelling wave-based and drift cell ion mobility mass spectrometry. Analytical Chemistry 81:248-254.
Thalassinos, K., Grabenauer, M., Slade, S. E., Hilton, G. R., Bowers, M. T. and Scrivens, J. H. (2009) Characterisation of phosphorylated peptides using travelling wave-based and drift cell ion mobility mass spectrometry. Analytical Chemistry 81:248-254.