Probing the structure of proteins
Ion Mobility Mass Spectrometry
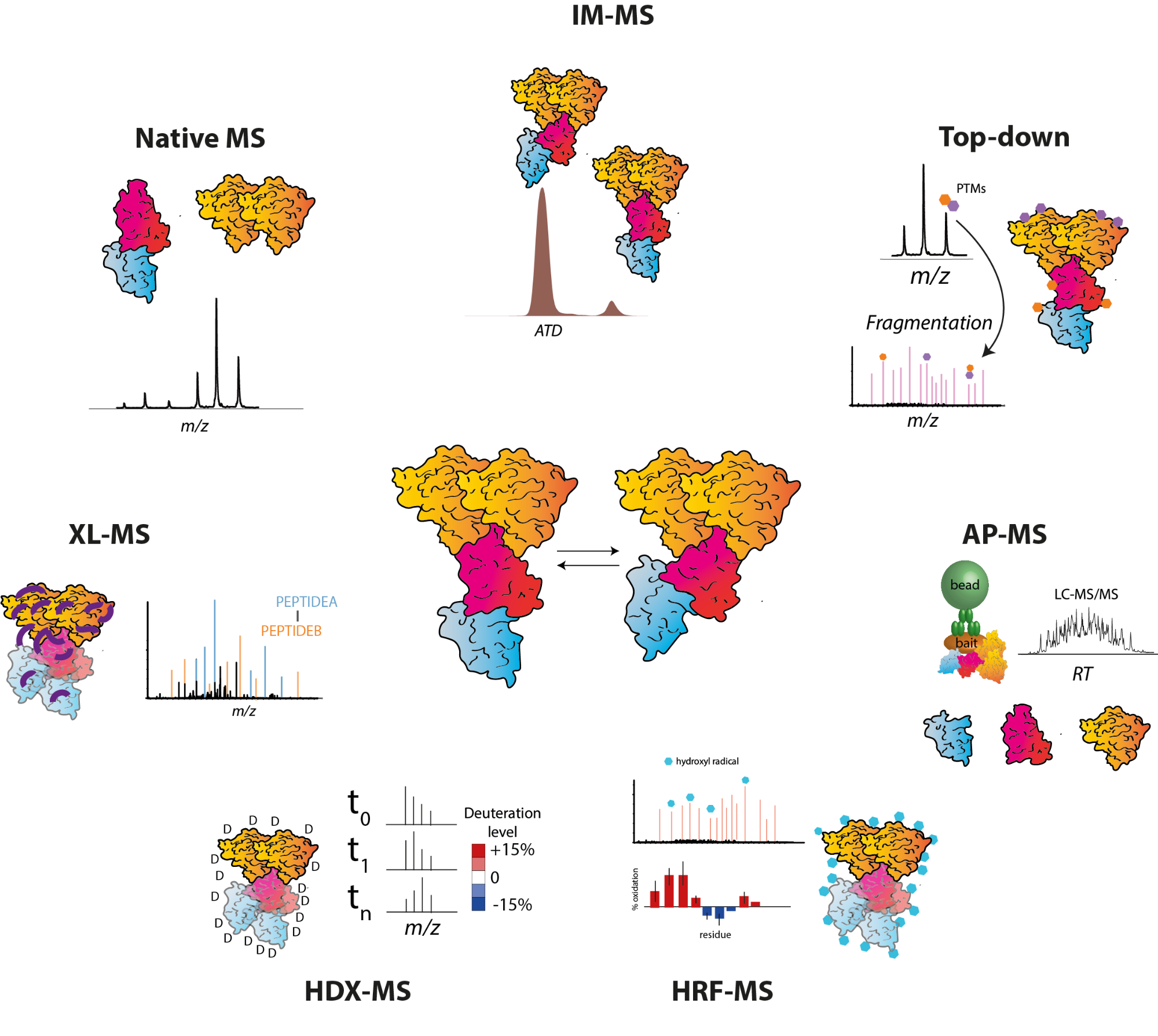
Modern protein prediction methods using deep neural networks can now achieve extremely accurate results when predicting protein structures from the protein's sequence. Despite this impressive achievement these approaches still struggle to predict the structure of flexible proteins, also known as intrinsically disordered proteins (IDPs) which comprise 30% of the human proteome, and those where a single folded state does not represent the majority conformation. They have also not so far tackled large multiprotein complexes which are often the major players within the cell. By contrast, structural mass spectrometry approaches (MS), and in particular ion mobility-mass spectrometry and crosslinking mass spectrometry, can probe the dynamic conformational landscape of proteins and proteins in complex with other molecules. In addition to in vitro studies, these approaches have recently been used to study the structure and dynamics of proteins on a proteome-wide scale and in their cellular environment. Combined with more established quantitative proteomics workflows, these analyses hold great promise in revealing the true behaviour and interactions of proteins within cells.
References
Behavior in the Gas Phase. Eldrid, C., Ben-Younis, A., Ujma, J., Britt, H., Cragnolini, T., Kalfas, S., Cooper-Shepherd, D., Tomczyk, N., Giles, K., Morris, M., Akter, R., Raleigh, D., Thalassinos, K. J Am Soc Mass Spectrom (2021) 32, 1545-1552
Gas Phase Stability of Protein Ions in a Cyclic Ion Mobility Spectrometry Traveling Wave Device. Eldrid, C., Ujma, J., Kalfas, S., Tomczyk, N., Giles, K., Morris, M., Thalassinos, K. Analytical Chemistry (2019) 91, 7554-7561
Developments in tandem ion mobility mass spectrometry Eldrid, C., Thalassinos, K. Biochem Soc Trans (2020) 48, 2457-2466