Software
I am involved in a number of software projects, most of which are built on top of the R platform for statistical computing, which is a free and open source programming language and environment comparable to Matlab, but with particular emphasis on statistics. Further information on the uses of R in neuroimaging can be found in a recent paper.
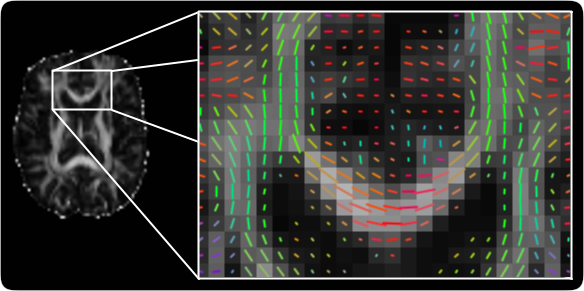
TractoR

The TractoR (for Tractography with R) project provides an R-based platform for magnetic resonance image processing and analysis, with a particular focus on diffusion-weighted imaging. As well as providing tools and R libraries for reading, manipulating and processing magnetic resonance images, the package offers algorithms for automated white matter segmentation and group analysis, including reference implementations of neighbourhood tractography methods.
The package is aimed at academic researchers working with magnetic resonance data of any sort, and is freely available under the terms of the GNU Public Licence, version 2. It is stable, contains a set of self-tests, and is developed according to good software engineering practice. Development is open, with full source code available. Substantial documentation is also provided.
Further information and downloads can be found via the project’s own web site. There is also an open-access paper published in the Journal of Statistical Software, which gives some technical details of the package. If you use TractoR in your publications, we would appreciate it if you cite this paper.
Other R packages
NiftyReg is a lightweight suite of image registration tools developed primarily by Marc Modat, Pankaj Daga and Sebastien Ourselin at UCL’s Centre for Medical Image Computing. It is part of the broader NifTK platform. I developed RNiftyReg, a package providing a clean and direct interface between NiftyReg and R. The package is available on CRAN, the Comprehensive R Archive Network, which is the biggest network for distribution of R packages.
The mmand package (for Mathematical Morphology in Any Number of Dimensions) can perform mathematical morphology and kernel operations such as smoothing on array-like data, such as images. It is focussed around a small number of flexible functions which can be used for a wide array of purposes—the package even includes an example of using them to simulate a cellular automaton.
SOMA (for Self-Organising Migrating Algorithm) is a general-purpose stochastic optimisation algorithm. The approach is similar to that of genetic algorithms, although it is based on the idea of a series of “migrations” by a fixed set of individuals, rather than the development of successive generations. It can be applied to any cost-minimisation problem with a bounded parameter space, and is robust to local minima. I have used it in my work on active imaging, and found its performance to be very good. I therefore put together an R implementation, available as the soma package, which is on CRAN.
R’s regular expression support is usable but not particularly fast or intuitive. I created the ore package as an alternative, which provides an R interface to the Onigmo C library. The package’s functions are consistent and fast, and offer additional features such as substitution functions, string escaping and a pattern dictionary.
Finally, the reportr package provides a general and flexible message reporting system for R. It is also available via CRAN.
Copyright © 2025 Jon Clayden. Any views or opinions expressed on this site are my own and not those of UCL.