R Introduction for UCL PhDs
UCL 2013
Florian Oswald. email me at f.oswald@ucl.ac.uk
Welcome to R!
Contents
- Some basic R. Not (at all) exaustive.
- These slides are designed to get you started.
- Try this online R challenge at codeschool. Codeschool provides excellent tryouts IN YOUR BROWSER (you don’t need to install anything.
- Have a look at many more resources about learning R on my UCL wiki
- Some quizzes
- Some example usage. We’ll be scratching the surface.
- A sample applied econometric project using the FES: reading, merging, analysing, transforming data, plots and regressions
- R and foreign Languages
- How to make this set of slides.
Why R?
- Licensing
- Availability of add on software (packages)
- Very easy to adapt to your needs. You can look at existing code, change it and reuse it.
- Ease of connecting to other (faster) software like fortran or C
- Who uses R? (Apart from Roger Koenker?)
Why not R?
- steep learning curve.
- “R is slow”. Although this is a very imprecise statement. See “calling C and Fortran”.
- the sheer extent of packages available makes is hard at time to find the most suitable one
- Some people lament that the CRAN model led to a bastardization of the language, and that it should be rewritten from scratch.
Learning R if you know Matlab/Stata/other
Supplementary material
- We’ll use some sample FES data and a Fortran function at some point
- if you are a git user type in your terminal
git clone git://github.com/floswald/R-demo.git
Using this set of slides
- hit
c on your keyboard for a linked table of contents. c again makes it disappear.
- left/right or mouse click or swipe takes you around
- code looks like this:
x <- 3.4
x # print x to screen
## [1] 3.4
- the first block is input, the second block (marked with
##) is output
- everything after
# is a comment
- you can (and should) type or copy and paste the code into your R and try around. All of the code you’ll see actually works.
- let’s do it.
Getting help()
- if you want help about a function, type
help(fname)
- if you don’t know which function you are looking for, try `??fname``
- go to the R Site Search
Basics
- Case-sensitive: x is different from X
- fully-fledged programming language. all the usual flow controls
- R needs to have all objects in memory (there are exeptions)
- R’s memory is more like matlabs and less like stata’s
- you can have many objects of different size, type etc
- (stata’s memory is one big spreadsheet. you can store a vector on the side, but it’s complicated.)
Assigning Values to Objects
- Everything in R (numbers, letters, vectors, functions, plots, formulas, calls,…) is an object
- assign a value (a number, a matrix, a word, a function, …) to an object with
<- or = . Use <- .
x <- 3.4
x
## [1] 3.4
reassigning overwrites the current value:
x <- "now x is a character string."
x
## [1] "now x is a character string."
x <- c(3, 5, 2.1, 1001, 4.6) # c() function combines single values into a vector.
x
## [1] 3.0 5.0 2.1 1001.0 4.6
The functions c(), str(x) and typeof(x)
typeof(x) tells you how x was stored, mode(x) is similar- among possible storage types are
logical: TRUE or FALSEinteger: 1, 2, 3, -10, …double: 1.0, pi, -3.2331, …factor: categorial variables
- dummy vars: (0,1) labelled
male,female
- categories: (1,2,3) labelled
(2,5],(5,7],(7,10]
complex: 0+0icharacter: “hello world”
c() requires values to have the same type (or it coerces into a common type)
typeof(x)
## [1] "double"
y <- letters[10:20] # letters number 10 trough 20 of the english alphabet
typeof(y)
## [1] "character"
typeof(c(x, y))
## [1] "character"
str(c(x, y)) # compact display of structure of an object
## chr [1:16] "3" "5" "2.1" "1001" "4.6" "j" "k" "l" ...
Subsetting a vector
- access the n-th element of
x with x[n]
- access any tuple of elements by suppling the indices
x <- rnorm(n = 8) # draw 8 random normal values
x
## [1] 0.4319 -0.9394 -0.2802 -0.8081 1.1112 0.1112 -0.5223 -0.6709
x[3:6] # '3:6' produces c(3,4,5,6)
## [1] -0.2802 -0.8081 1.1112 0.1112
x[c(1, 5, 8)] # elts 1,5 and 8
## [1] 0.4319 1.1112 -0.6709
x[-c(1, 5, 8)] # all elts except 1,5 and 8
## [1] -0.9394 -0.2802 -0.8081 0.1112 -0.5223
Helper Functions for allocation: seq() and rep()
x <- seq(from = 1, to = 15, by = 3) # also 'length' instead of 'by'. like matlab linspace()
x
## [1] 1 4 7 10 13
str(x)
## num [1:5] 1 4 7 10 13
y <- rep(1:3, c(2, 3, 4))
y
## [1] 1 1 2 2 2 3 3 3 3
z <- rep(c("oh my word"), 3)
z
## [1] "oh my word" "oh my word" "oh my word"
Your turn 1
- create a vector x with values 0, 3, 6, 9, 12, 15, 18
- what is the
typeof(x)?
- create a vector y containing the words {“are”,“my”,“favourite”,“numbers”}
- create a vector z from the last 2 elements of x and all of y
- what’s the
typeof(z)?
Your solution 1
x <- seq(from = 0, to = 18, by = 3)
x
## [1] 0 3 6 9 12 15 18
typeof(x)
## [1] "double"
y <- c("are", "my", "favourite", "numbers")
z <- c(x[c(6, 7)], y) # R 'coerces' x and y to a common type. here: char
z
## [1] "15" "18" "are" "my" "favourite" "numbers"
typeof(z)
## [1] "character"
Some Vector Arithmetic
- Operations +-/* are element by element
x <- 1:3
y <- 4:6
x + y
## [1] 5 7 9
- and vectors are “recycled” if they are not of the same length
x <- 1:4
x + y # recycling the shorter vector y. R gives a warning.
## Warning: longer object length is not a multiple of shorter object length
## [1] 5 7 9 8
matrix
m1 <- matrix(data = 1:9, nrow = 3, ncol = 3, byrow = TRUE)
m2 <- matrix(data = 1:12, nrow = 4, ncol = 3, byrow = FALSE)
m1
## [,1] [,2] [,3]
## [1,] 1 2 3
## [2,] 4 5 6
## [3,] 7 8 9
m2
## [,1] [,2] [,3]
## [1,] 1 5 9
## [2,] 2 6 10
## [3,] 3 7 11
## [4,] 4 8 12
Combining matrices
- similarly to
c() for vectors, we have cbind() and rbind()
- column-bind and row-bind
rbind(m1, m2) # glue together the last row of of m1 and first of m2
## [,1] [,2] [,3]
## [1,] 1 2 3
## [2,] 4 5 6
## [3,] 7 8 9
## [4,] 1 5 9
## [5,] 2 6 10
## [6,] 3 7 11
## [7,] 4 8 12
cbind(m1, t(m2)) # glue last col of m1 and first of t(m2)
## [,1] [,2] [,3] [,4] [,5] [,6] [,7]
## [1,] 1 2 3 1 2 3 4
## [2,] 4 5 6 5 6 7 8
## [3,] 7 8 9 9 10 11 12
Subsetting a matrix
- get size of matrix with
dim(m1)
- you can select indices of rows and columns. Very similar to matlab.
m1[2, ] # row 2, all columns
## [1] 4 5 6
m2[, 1] # all rows, column 1
## [1] 1 2 3 4
m1[c(1, 3), c(2, 3)] # rows 1,3 and cols 2,3
## [,1] [,2]
## [1,] 2 3
## [2,] 8 9
- or by name, if those were given:
colnames(m1) <- c("col1", "col2", "col3")
m1[, "col2"]
## [1] 2 5 8
colnames(m1) <- NULL # remove colnames
basic matrix arithmetic
Again, +-/* are element-wise. Works only on equal sized matrices:
m1 + m2 # error
## Error: non-conformable arrays
m1 * m2 # error
## Error: non-conformable arrays
%*% is the matrix muliplication operation. matrices need to be conformable:
m2 %*% m1
## [,1] [,2] [,3]
## [1,] 84 99 114
## [2,] 96 114 132
## [3,] 108 129 150
## [4,] 120 144 168
m1 %*% t(m2)
## [,1] [,2] [,3] [,4]
## [1,] 38 44 50 56
## [2,] 83 98 113 128
## [3,] 128 152 176 200
m1 %*% m2
## Error: non-conformable arguments
data.frame
- A data.frame is a standard dataset
- very similar to
matrix
- each row is an observation, each column is a variable
- create a data.frame from scratch with
data.frame()
- Notice: all columns need to be of the same length (or recyclable)! (a data.frame is a rectangle; it’s a spreadsheet!)
df <- data.frame(cat.1 = rep(1:3, each = 2), cat.2 = 1:2, values = rnorm(6))
dim(df)
## [1] 6 3
df
## cat.1 cat.2 values
## 1 1 1 0.8164
## 2 1 2 0.6714
## 3 2 1 -0.2564
## 4 2 2 -0.8258
## 5 3 1 -0.7887
## 6 3 2 -0.6103
Subsetting a data.frame
- A data.frame is similar to a matrix. Subsetting is thus similar.
- but additionally, you can access columns with the
$ operator:
df$values
## [1] 0.8164 0.6714 -0.2564 -0.8258 -0.7887 -0.6103
- the
$ in general accesses elements of “lists” (see below)
- if an element does not exist,
$ adds it:
df$new.col <- df$cat.1 + df$values
df
## cat.1 cat.2 values new.col
## 1 1 1 0.8164 1.816
## 2 1 2 0.6714 1.671
## 3 2 1 -0.2564 1.744
## 4 2 2 -0.8258 1.174
## 5 3 1 -0.7887 2.211
## 6 3 2 -0.6103 2.390
subset() 1
- get a subset of observations conditional on values of a variable
subset(df, cat.1 == 1) # select all obs were cat.1 equals 1
## cat.1 cat.2 values new.col
## 1 1 1 0.8164 1.816
## 2 1 2 0.6714 1.671
- this is the same as
df[df$cat.1==1,]
subset() 2
- get a subset of variables
subset(df, select = c(cat.2, values)) # selects columns 'cat.2' and 'values'
## cat.2 values
## 1 1 0.8164
## 2 2 0.6714
## 3 1 -0.2564
## 4 2 -0.8258
## 5 1 -0.7887
## 6 2 -0.6103
Removing columns
remove a column with NULL:
df$new.col <- NULL
df
## cat.1 cat.2 values
## 1 1 1 0.8164
## 2 1 2 0.6714
## 3 2 1 -0.2564
## 4 2 2 -0.8258
## 5 3 1 -0.7887
## 6 3 2 -0.6103
Appending Data: rbind
- very similar to
rbind for matrix
- If you have two data.frames with equally named columns (e.g. two editions of the same dataset), just glue together with
rbind
df2 <- df # create df2 as an exact copy of df
df2$values <- 1:nrow(df) # but change the entries for 'values'
rbind(df, df2) # join them row-wise
## cat.1 cat.2 values
## 1 1 1 0.8164
## 2 1 2 0.6714
## 3 2 1 -0.2564
## 4 2 2 -0.8258
## 5 3 1 -0.7887
## 6 3 2 -0.6103
## 7 1 1 1.0000
## 8 1 2 2.0000
## 9 2 1 3.0000
## 10 2 2 4.0000
## 11 3 1 5.0000
## 12 3 2 6.0000
Looking at data.frames
- R stores datasets typically as data.frames
- special “methods”" to describe those (a method is a function that does things differently depending on the object you give it)
data(LifeCycleSavings) # data() shows available built-in datasets
LS <- LifeCycleSavings
head(LS) # show the first 6 rows of data.frame LifecycleSavings.
## sr pop15 pop75 dpi ddpi
## Australia 11.43 29.35 2.87 2329.7 2.87
## Austria 12.07 23.32 4.41 1508.0 3.93
## Belgium 13.17 23.80 4.43 2108.5 3.82
## Bolivia 5.75 41.89 1.67 189.1 0.22
## Brazil 12.88 42.19 0.83 728.5 4.56
## Canada 8.79 31.72 2.85 2982.9 2.43
summary(LS) # function summary()
## sr pop15 pop75 dpi
## Min. : 0.60 Min. :21.4 Min. :0.56 Min. : 89
## 1st Qu.: 6.97 1st Qu.:26.2 1st Qu.:1.12 1st Qu.: 288
## Median :10.51 Median :32.6 Median :2.17 Median : 696
## Mean : 9.67 Mean :35.1 Mean :2.29 Mean :1107
## 3rd Qu.:12.62 3rd Qu.:44.1 3rd Qu.:3.33 3rd Qu.:1796
## Max. :21.10 Max. :47.6 Max. :4.70 Max. :4002
## ddpi
## Min. : 0.22
## 1st Qu.: 2.00
## Median : 3.00
## Mean : 3.76
## 3rd Qu.: 4.48
## Max. :16.71
ordering a data.frame
- rearrange row indices by a certain criterion
order() and sort()- function order(x) returns the indices of x in ascending/descending order:
order(c(5, 2, 6, 1), decreasing = TRUE)
## [1] 3 1 2 4
- plugging in a vector of indices into a matrix/data.frame, reorders the matrix according to those:
save.ranking <- order(LS$sr, decreasing = TRUE)
head(LS[save.ranking, ])
## sr pop15 pop75 dpi ddpi
## Japan 21.10 27.01 1.91 1257.3 8.21
## Zambia 18.56 45.25 0.56 138.3 5.14
## Denmark 16.85 24.42 3.93 2496.5 3.99
## Malta 15.48 32.54 2.47 601.0 8.12
## Netherlands 14.65 24.71 3.25 1740.7 7.66
## Italy 14.28 24.52 3.48 1390.0 3.54
packages
- “packages” are add on libraries, i.e. collections of functions, algorithms and datasets
- The great number of available packages is one of the strengths of R.
- packages on CRAN have a certain degree of quality control
installing packages
- install a packages available at CRAN simply by typing, for example
install.packages("ggplot2")
ggplot2 is a widely used package with plotting functions.- load a package by calling the library function:
library(ggplot2) # now the content of ggplot2 (functions, data, etc) are 'visible'
cut_interval # for example, here's the code to function 'cut_interval'
## function (x, n = NULL, length = NULL, ...)
## {
## cut(x, breaks(x, "width", n, length), include.lowest = TRUE,
## ...)
## }
## <environment: namespace:ggplot2>
- get help for a package via
help(package="package-name")
Functions
- R relies heavily on functions
- in fact, R is composed of functions
- look at the source code of any R function by typing the name:
matrix
## function (data = NA, nrow = 1, ncol = 1, byrow = FALSE, dimnames = NULL)
## {
## if (is.object(data) || !is.atomic(data))
## data <- as.vector(data)
## .Internal(matrix(data, nrow, ncol, byrow, dimnames, missing(nrow),
## missing(ncol)))
## }
## <bytecode: 0x100c40320>
## <environment: namespace:base>
- you can of course define your own functions.
- for example, a function to compute the interval that is “spanned” by a vector, i.e. the interval between the min and max of vector:
span <- function(x) {
stopifnot(is.numeric(x)) # stops if x is not numeric
r <- range(x) # range() gives the range
rval <- abs(r[2] - r[1]) # computes and returns the interval spanned by x
return(rval) # returns result
}
myvec <- rnorm(50) # draws 50 values from the standard normal pdf
span(myvec)
## [1] 4.163
Lists
- can contain data in any “mode”: vectors, matrices, data.frames, characters, functions, plots, and more lists
- similar to a
matlab or C structure
l <- list(words = c("oh my word(s)"), mats = list(mat1 = m1, mat2 = m2),
funs = span)
l
## $words
## [1] "oh my word(s)"
##
## $mats
## $mats$mat1
## [,1] [,2] [,3]
## [1,] 1 2 3
## [2,] 4 5 6
## [3,] 7 8 9
##
## $mats$mat2
## [,1] [,2] [,3]
## [1,] 1 5 9
## [2,] 2 6 10
## [3,] 3 7 11
## [4,] 4 8 12
##
##
## $funs
## function (x)
## {
## stopifnot(is.numeric(x))
## r <- range(x)
## rval <- abs(r[2] - r[1])
## return(rval)
## }
Working with Lists
- Access a list element by position number in the list, or by name (if given)
[["x"]] gives you element “x” as a standalone object["x"] gives you element “x” as a sublist with element “x”
l[[2]] # second list element: 'mats'
## $mat1
## [,1] [,2] [,3]
## [1,] 1 2 3
## [2,] 4 5 6
## [3,] 7 8 9
##
## $mat2
## [,1] [,2] [,3]
## [1,] 1 5 9
## [2,] 2 6 10
## [3,] 3 7 11
## [4,] 4 8 12
l$mats$mat1 # of that element, the first element
## [,1] [,2] [,3]
## [1,] 1 2 3
## [2,] 4 5 6
## [3,] 7 8 9
str(l[[1]]) # extract first element completely
## chr "oh my word(s)"
str(l[1]) # extract first element as member of sublist
## List of 1
## $ words: chr "oh my word(s)"
Adding and removing a new elements from a list
l$new.element <- rnorm(5) # add a new element: 5 random draws from the standard normal
l$bool.value <- l$new.element > 0 # add a new element: previously drawn numbers positive?
l$new.element <- NULL # delete an element
l$bool.value <- NULL
l$words <- NULL
l$funs <- NULL
l
## $mats
## $mats$mat1
## [,1] [,2] [,3]
## [1,] 1 2 3
## [2,] 4 5 6
## [3,] 7 8 9
##
## $mats$mat2
## [,1] [,2] [,3]
## [1,] 1 5 9
## [2,] 2 6 10
## [3,] 3 7 11
## [4,] 4 8 12
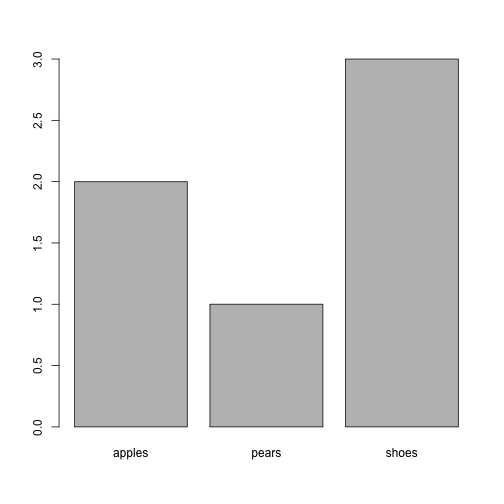
Your Turn 2
- make a list “fruit” with two elements: “apples” and “pears”.
- for both apples and pears make up 3 pairs of random price/quantity values, e.g. for apples
- for both apples and pears, store those values (i.e. the above table) in a data.frame with colum names “price” and “quantity”
- In other words, produce the following list. (make up your own numbers)
## $apples
## price quantity
## 1 3.594 4
## 2 2.632 1
## 3 1.772 3
##
## $pears
## price quantity
## 1 1.748 1
## 2 3.635 4
## 3 2.773 2
Your Solution 2
- you can input the values by hand, for example
apples <- data.frame(price=c(1.1,2.9,1.6),quantity=c(2,3,1))
- I used the uniform random generator
runif() for prices and sample() for quantities
apples <- data.frame(price = runif(min = 0, max = 4, n = 3), quantity = sample(1:4,
size = 3))
pears <- data.frame(price = runif(min = 0, max = 4, n = 3), quantity = sample(1:4,
size = 3))
fruit <- list()
fruit$apples <- apples
fruit$pears <- pears
# or all in one line:
fruit <- list(apples = data.frame(price = runif(min = 0, max = 4,
n = 3), quantity = sample(1:4, size = 3)), pears = data.frame(price = runif(min = 0,
max = 4, n = 3), quantity = sample(1:4, size = 3)))
Factors
- Factors are categorical variables
- a number is given a character label
- R has plotting and tabulating methods for factors
- also nice useage in regressions
new.fac <- factor(x = c(1, 1, 2, 5, 5, 5), labels = c("apples", "pears",
"shoes"))
new.fac
## [1] apples apples pears shoes shoes shoes
## Levels: apples pears shoes
table(new.fac)
## new.fac
## apples pears shoes
## 2 1 3
plot(new.fac)
Contingency Tables of multiple Factors
table() can count cases of factors against each other
UCB <- as.data.frame(UCBAdmissions) # load built-in data on Berkeley admissions
tail(UCB) # this time, display the last 6 rows
## Admit Gender Dept Freq
## 19 Admitted Female E 94
## 20 Rejected Female E 299
## 21 Admitted Male F 22
## 22 Rejected Male F 351
## 23 Admitted Female F 24
## 24 Rejected Female F 317
summary(UCB) # notice how summary treats factors and numerics
## Admit Gender Dept Freq
## Admitted:12 Male :12 A:4 Min. : 8
## Rejected:12 Female:12 B:4 1st Qu.: 80
## C:4 Median :170
## D:4 Mean :189
## E:4 3rd Qu.:302
## F:4 Max. :512
table(UCB$Admit, UCB$Dept) # table admit vs Dept?
##
## A B C D E F
## Admitted 2 2 2 2 2 2
## Rejected 2 2 2 2 2 2
- But is that what we want here?
Contingency tables 2
- No! We want to know “Freq” by those categories (those
factors).
- In R-speak, what is the relationship
Freq ~ factors?
xtabs()- takes a
formula as an argument
- typical formula:
y ~ x + z means “y is explained by x and z”
xtabs(Freq ~ Admit + Dept, data = UCB) # Freq is explained by Admit and Dept
## Dept
## Admit A B C D E F
## Admitted 601 370 322 269 147 46
## Rejected 332 215 596 523 437 668
xtabs(Freq ~ Admit + Gender, data = UCB)
## Gender
## Admit Male Female
## Admitted 1198 557
## Rejected 1493 1278
Your turn 3
- create a table that shows the number of admissions/rejections by gender AND department.
Your Solution 3
xtabs(Freq ~ Admit + Gender + Dept, data = UCB)
## , , Dept = A
##
## Gender
## Admit Male Female
## Admitted 512 89
## Rejected 313 19
##
## , , Dept = B
##
## Gender
## Admit Male Female
## Admitted 353 17
## Rejected 207 8
##
## , , Dept = C
##
## Gender
## Admit Male Female
## Admitted 120 202
## Rejected 205 391
##
## , , Dept = D
##
## Gender
## Admit Male Female
## Admitted 138 131
## Rejected 279 244
##
## , , Dept = E
##
## Gender
## Admit Male Female
## Admitted 53 94
## Rejected 138 299
##
## , , Dept = F
##
## Gender
## Admit Male Female
## Admitted 22 24
## Rejected 351 317
Workspace
- In general, R needs to load an object into memory to be able to work with it
- This can be a problem with large data sets
- the function
ls() shows the content of your workspace, or your current “search path”
ls()
## [1] "%ni%" "apples" "df"
## [4] "df2" "fruit" "l"
## [7] "LifeCycleSavings" "LS" "m1"
## [10] "m2" "myvec" "new.fac"
## [13] "pears" "save.ranking" "span"
## [16] "UCB" "x" "y"
## [19] "z"
rm(df, m1, m2, "%ni%", fruit, l) # remove objects
ls()
## [1] "apples" "df2" "LifeCycleSavings"
## [4] "LS" "myvec" "new.fac"
## [7] "pears" "save.ranking" "span"
## [10] "UCB" "x" "y"
## [13] "z"
rm(list = ls(all = TRUE)) # remove all objects
ls()
## character(0)
A Sample Applied Econometric Project
Food and Expenditure Survey (FES)
- the repository you downloaded from github contains a folder “data”
- contains 4 files, 2 in .csv and 2 in .dta format
- each format has one data file (“fesdat”) and one file on income (“fesinc”)
Agenda
- how to read different data
- how to merge data
- how to analyse data
- with summary statistics (numerical variables)
- in tables (categorical vars)
- graphically
- with statistical models
- how to report results
R does a lot of Econometrics
Reading Data Files
- Read data with with family of
read.xxx() functions:
read.csv(): comma separated valuesread.dta(): stata formatread.table(): generic text/table readerlibrary(foreign) contains many useful functions to read all sorts of foreign data formatslibrary(xlsx) is useful to read .xls data
Read FES
- let’s read both .csv and .dta formats just to demonstrate:
setwd(dir = "~/git/R-demo/") # set working directory
fesdat.csv <- read.csv(file = "data/fesdat.csv") # read the data in csv format. Note that 'file' could also be a URL
fesinc.csv <- read.csv(file = "data/fesinc.csv") # read the income data in csv format
library(foreign) # load foreign to read stata data
fesdat.dta <- read.dta(file = "data/fesdat2005.dta")
fesinc.dta <- read.dta(file = "data/fesinc2005.dta")
head(fesdat.dta) # do they look the same?
## hhref year numads numadret numadern numhhkid kids0 kids1 kids2 kids34
## 1 2478 2005 2 0 2 0 0 0 0 0
## 2 53 2005 1 0 0 1 0 0 1 0
## 3 1085 2005 1 0 0 1 0 0 1 0
## 4 6399 2005 1 0 0 1 1 0 0 0
## 5 2441 2005 1 0 0 1 1 0 0 0
## 6 2431 2005 1 0 0 1 1 0 0 0
## kids510 kids1116 kids1718 ncars nrooms age sex
## 1 0 0 0 1 4 17 2
## 2 0 0 0 0 5 18 2
## 3 0 0 0 0 5 18 2
## 4 0 0 0 0 5 18 2
## 5 0 0 0 0 5 18 2
## 6 0 0 0 0 4 18 2
## marstat ageced educ hours foodin foodout
## 1 cohabiting 16 0 40 38.12 15.845
## 2 Single,nev marr inc wid,div,sep pre 91 0 8 0 16.41 65.635
## 3 Single,nev marr inc wid,div,sep pre 91 16 0 0 21.39 3.300
## 4 Single,nev marr inc wid,div,sep pre 91 16 0 0 30.11 9.265
## 5 Single,nev marr inc wid,div,sep pre 91 17 0 0 8.89 5.170
## 6 Single,nev marr inc wid,div,sep pre 91 16 0 0 15.74 19.620
## alc nondur semidur durables wfoodin wfoodout walc ndex lndex
## 1 8.64 171.40 11.285 11.70 0.2087 0.08673 0.04729 182.69 5.208
## 2 3.00 97.51 23.490 0.00 0.1356 0.54244 0.02479 121.00 4.796
## 3 20.00 110.33 7.500 0.00 0.1815 0.02801 0.16974 117.83 4.769
## 4 0.00 79.78 0.875 3.85 0.3733 0.11488 0.00000 80.65 4.390
## 5 0.00 34.38 26.480 2.50 0.1461 0.08495 0.00000 60.86 4.109
## 6 9.32 103.85 14.500 71.54 0.1330 0.16577 0.07875 118.35 4.774
head(fesdat.csv)
## hhref year numads numadret numadern numhhkid kids0 kids1 kids2 kids34
## 1 2478 2005 2 0 2 0 0 0 0 0
## 2 53 2005 1 0 0 1 0 0 1 0
## 3 1085 2005 1 0 0 1 0 0 1 0
## 4 6399 2005 1 0 0 1 1 0 0 0
## 5 2441 2005 1 0 0 1 1 0 0 0
## 6 2431 2005 1 0 0 1 1 0 0 0
## kids510 kids1116 kids1718 ncars nrooms age sex
## 1 0 0 0 1 4 17 2
## 2 0 0 0 0 5 18 2
## 3 0 0 0 0 5 18 2
## 4 0 0 0 0 5 18 2
## 5 0 0 0 0 5 18 2
## 6 0 0 0 0 4 18 2
## marstat ageced educ hours foodin foodout
## 1 cohabiting 16 0 40 38.12 15.845
## 2 Single,nev marr inc wid,div,sep pre 91 0 8 0 16.41 65.635
## 3 Single,nev marr inc wid,div,sep pre 91 16 0 0 21.39 3.300
## 4 Single,nev marr inc wid,div,sep pre 91 16 0 0 30.11 9.265
## 5 Single,nev marr inc wid,div,sep pre 91 17 0 0 8.89 5.170
## 6 Single,nev marr inc wid,div,sep pre 91 16 0 0 15.74 19.620
## alc nondur semidur durables wfoodin wfoodout walc ndex lndex
## 1 8.64 171.40 11.285 11.70 0.2087 0.08673 0.04729 182.69 5.208
## 2 3.00 97.51 23.490 0.00 0.1356 0.54244 0.02479 121.00 4.796
## 3 20.00 110.33 7.500 0.00 0.1815 0.02801 0.16974 117.83 4.769
## 4 0.00 79.78 0.875 3.85 0.3733 0.11488 0.00000 80.65 4.390
## 5 0.00 34.38 26.480 2.50 0.1461 0.08495 0.00000 60.86 4.109
## 6 9.32 103.85 14.500 71.54 0.1330 0.16577 0.07875 118.35 4.774
Summary
dat <- fesdat.dta # let's rename this to something shorter
inc <- fesinc.dta
summary(dat)
## hhref year numads numadret
## Min. : 1 Min. :2005 Min. :1.0 Min. :0.000
## 1st Qu.:1697 1st Qu.:2005 1st Qu.:1.0 1st Qu.:0.000
## Median :3393 Median :2005 Median :2.0 Median :0.000
## Mean :3393 Mean :2005 Mean :1.8 Mean :0.423
## 3rd Qu.:5089 3rd Qu.:2005 3rd Qu.:2.0 3rd Qu.:1.000
## Max. :6785 Max. :2005 Max. :9.0 Max. :3.000
##
## numadern numhhkid kids0 kids1
## Min. :0.00 Min. :0.000 Min. :0.0000 Min. :0.0000
## 1st Qu.:0.00 1st Qu.:0.000 1st Qu.:0.0000 1st Qu.:0.0000
## Median :1.00 Median :0.000 Median :0.0000 Median :0.0000
## Mean :1.05 Mean :0.573 Mean :0.0292 Mean :0.0283
## 3rd Qu.:2.00 3rd Qu.:1.000 3rd Qu.:0.0000 3rd Qu.:0.0000
## Max. :9.00 Max. :8.000 Max. :2.0000 Max. :2.0000
##
## kids2 kids34 kids510 kids1116
## Min. :0.0000 Min. :0.000 Min. :0.000 Min. :0.00
## 1st Qu.:0.0000 1st Qu.:0.000 1st Qu.:0.000 1st Qu.:0.00
## Median :0.0000 Median :0.000 Median :0.000 Median :0.00
## Mean :0.0315 Mean :0.059 Mean :0.199 Mean :0.19
## 3rd Qu.:0.0000 3rd Qu.:0.000 3rd Qu.:0.000 3rd Qu.:0.00
## Max. :2.0000 Max. :5.000 Max. :4.000 Max. :4.00
##
## kids1718 ncars nrooms age
## Min. :0.0000 Min. :0.00 Min. :1.00 Min. :17.0
## 1st Qu.:0.0000 1st Qu.:0.00 1st Qu.:4.00 1st Qu.:38.0
## Median :0.0000 Median :1.00 Median :5.00 Median :51.0
## Mean :0.0361 Mean :1.08 Mean :5.11 Mean :51.9
## 3rd Qu.:0.0000 3rd Qu.:2.00 3rd Qu.:6.00 3rd Qu.:66.0
## Max. :2.0000 Max. :7.00 Max. :6.00 Max. :80.0
##
## sex marstat
## Min. :1.00 Married spouse in household :3349
## 1st Qu.:1.00 Single,nev marr inc wid,div,sep pre 91:1008
## Median :1.00 Widowed : 833
## Mean :1.42 Divorced : 722
## 3rd Qu.:2.00 cohabiting : 616
## Max. :2.00 Separated : 252
## (Other) : 5
## ageced educ hours foodin
## Min. : 0.0 Min. :0.000 Min. : 0.0 Min. : 0.0
## 1st Qu.:15.0 1st Qu.:0.000 1st Qu.: 0.0 1st Qu.: 24.0
## Median :16.0 Median :0.000 Median : 0.0 Median : 40.0
## Mean :16.8 Mean :0.117 Mean : 17.4 Mean : 45.4
## 3rd Qu.:18.0 3rd Qu.:0.000 3rd Qu.: 38.0 3rd Qu.: 60.9
## Max. :97.0 Max. :8.000 Max. :104.0 Max. :329.2
##
## foodout alc nondur semidur
## Min. : 0.0 Min. : 0.0 Min. : 0 Min. : 0.0
## 1st Qu.: 3.5 1st Qu.: 0.0 1st Qu.: 120 1st Qu.: 1.2
## Median : 13.0 Median : 6.0 Median : 203 Median : 15.0
## Mean : 21.0 Mean : 14.3 Mean : 257 Mean : 33.3
## 3rd Qu.: 29.5 3rd Qu.: 20.0 3rd Qu.: 322 3rd Qu.: 43.0
## Max. :390.2 Max. :274.4 Max. :10512 Max. :811.9
##
## durables wfoodin wfoodout walc
## Min. :-243.2 Min. :0.000 Min. :0.0000 Min. :0.0000
## 1st Qu.: 1.5 1st Qu.:0.117 1st Qu.:0.0201 1st Qu.:0.0000
## Median : 15.4 Median :0.184 Median :0.0565 Median :0.0250
## Mean : 55.6 Mean :0.202 Mean :0.0701 Mean :0.0498
## 3rd Qu.: 60.2 3rd Qu.:0.263 3rd Qu.:0.1033 3rd Qu.:0.0715
## Max. :2243.8 Max. :0.882 Max. :0.7741 Max. :0.6427
## NA's :1 NA's :1 NA's :1
## ndex lndex
## Min. : 0 Min. :0.837
## 1st Qu.: 131 1st Qu.:4.876
## Median : 229 Median :5.432
## Mean : 290 Mean :5.380
## 3rd Qu.: 368 3rd Qu.:5.908
## Max. :10533 Max. :9.262
## NA's :1
dat$year <- NULL # we've only got one year, so useless info
Factors
str(dat$marstat) # marstat is already a factor
## Factor w/ 8 levels "not recorded",..: 4 5 5 5 5 5 5 5 4 4 ...
levels(dat$marstat)
## [1] "not recorded"
## [2] "Married spouse in household"
## [3] "Married spouse not in household"
## [4] "cohabiting"
## [5] "Single,nev marr inc wid,div,sep pre 91"
## [6] "Widowed"
## [7] "Divorced"
## [8] "Separated"
dat$sex <- factor(dat$sex, labels = c("male", "female")) # convert sex into a factor
summary(subset(dat, select = c(sex, marstat)))
## sex marstat
## male :3958 Married spouse in household :3349
## female:2827 Single,nev marr inc wid,div,sep pre 91:1008
## Widowed : 833
## Divorced : 722
## cohabiting : 616
## Separated : 252
## (Other) : 5
Contingency Tables of Factors
attach(dat) # attach(x) makes all cols of x visible on the search path
table(sex) # otherwise I'd have to type dat$sex here
## sex
## male female
## 3958 2827
table(marstat, sex)
## sex
## marstat male female
## not recorded 0 0
## Married spouse in household 2546 803
## Married spouse not in household 4 1
## cohabiting 412 204
## Single,nev marr inc wid,div,sep pre 91 474 534
## Widowed 220 613
## Divorced 224 498
## Separated 78 174
More complicated tables: xtabs()
f <- ~sex + kids0
class(f) # formula with only RHS 'explaining' variables: just get a count
## [1] "formula"
xtabs(f)
## kids0
## sex 0 1 2
## male 3841 115 2
## female 2749 77 1
xtabs(~kids0 + sex + nrooms) # three way table
## , , nrooms = 1
##
## sex
## kids0 male female
## 0 8 5
## 1 0 0
## 2 0 0
##
## , , nrooms = 2
##
## sex
## kids0 male female
## 0 41 36
## 1 0 0
## 2 0 0
##
## , , nrooms = 3
##
## sex
## kids0 male female
## 0 232 226
## 1 7 2
## 2 0 0
##
## , , nrooms = 4
##
## sex
## kids0 male female
## 0 600 581
## 1 18 28
## 2 1 0
##
## , , nrooms = 5
##
## sex
## kids0 male female
## 0 931 797
## 1 30 20
## 2 0 1
##
## , , nrooms = 6
##
## sex
## kids0 male female
## 0 2029 1104
## 1 60 27
## 2 1 0
prop.table(xtabs(~sex + kids0), margin = 1) # proportion of kids0 by sex
## kids0
## sex 0 1 2
## male 0.9704396 0.0290551 0.0005053
## female 0.9724089 0.0272374 0.0003537
prop.table(xtabs(~sex + kids0), margin = 2) # proportion of kids0 within age
## kids0
## sex 0 1 2
## male 0.5829 0.5990 0.6667
## female 0.4171 0.4010 0.3333
detach(dat) # remove dat from search path
Merge Data
- Both
dat and inc include a unique hh identifier hhref
- use
merge()
dat.inc <- merge(dat, inc, by = c("hhref")) # if multiple keys: by=c('hhref','year')
head(dat.inc) # note new column hhinc
## hhref numads numadret numadern numhhkid kids0 kids1 kids2 kids34 kids510
## 1 1 1 1 0 0 0 0 0 0 0
## 2 2 2 0 2 1 0 0 0 0 0
## 3 3 1 0 0 0 0 0 0 0 0
## 4 4 2 1 1 0 0 0 0 0 0
## 5 5 3 0 3 1 0 0 0 0 0
## 6 6 1 0 1 0 0 0 0 0 0
## kids1116 kids1718 ncars nrooms age sex
## 1 0 0 1 6 66 female
## 2 1 0 0 4 38 male
## 3 0 0 0 3 25 male
## 4 0 0 0 3 80 female
## 5 1 0 2 6 40 male
## 6 0 0 1 4 50 female
## marstat ageced educ hours foodin foodout
## 1 Widowed 15 0 0 64.21 26.27
## 2 cohabiting 16 0 39 75.67 10.99
## 3 Single,nev marr inc wid,div,sep pre 91 0 6 0 11.86 6.67
## 4 Divorced 14 0 0 58.23 8.09
## 5 Married spouse in household 16 0 50 63.42 53.47
## 6 Divorced 15 0 5 19.73 5.32
## alc nondur semidur durables wfoodin wfoodout walc ndex lndex year
## 1 15.07 240.39 45.34 37.65 0.2247 0.09194 0.05274 285.73 5.655 2005
## 2 26.07 257.56 10.50 13.99 0.2823 0.04100 0.09727 268.06 5.591 2005
## 3 0.00 38.13 0.00 0.00 0.3110 0.17493 0.00000 38.13 3.641 2005
## 4 23.61 142.49 226.82 86.35 0.1577 0.02191 0.06393 369.32 5.912 2005
## 5 24.74 321.29 36.74 139.17 0.1771 0.14936 0.06910 358.02 5.881 2005
## 6 19.91 79.15 0.00 0.00 0.2492 0.06721 0.25148 79.15 4.371 2005
## hhinc
## 1 279.95
## 2 467.07
## 3 40.00
## 4 285.46
## 5 610.95
## 6 27.18
Create Bins: cut() an interval
- suppose instead of
age = {17,18,19,...} we want categories of age
- base R has
cut()
- ggplot2 has derived
cut_interval(), bit more intuitive
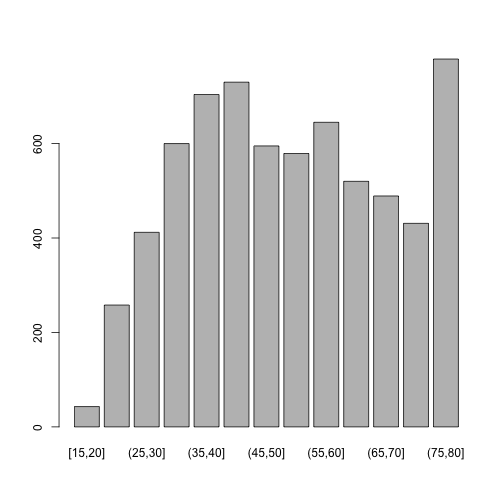
dat.inc$age.cat <- cut_interval(dat.inc$age, length = 5) # add col with age category
dat.inc$ced.cat <- cut_interval(dat.inc$ageced, length = 5) # add col with age at educ completion category
str(dat.inc$age.cat) # is a factor?
## Factor w/ 13 levels "[15,20]","(20,25]",..: 11 5 2 13 5 7 3 3 3 6 ...
table(dat.inc$age.cat)
##
## [15,20] (20,25] (25,30] (30,35] (35,40] (40,45] (45,50] (50,55] (55,60]
## 43 258 412 600 704 730 595 579 645
## (60,65] (65,70] (70,75] (75,80]
## 520 489 431 779
plot(dat.inc$age.cat)
table(dat.inc$ced.cat)
##
## [0,5] (5,10] (10,15] (15,20] (20,25] (25,30] (30,35] (35,40]
## 103 4 2201 3471 931 55 7 0
## (40,45] (45,50] (50,55] (55,60] (60,65] (65,70] (70,75] (75,80]
## 1 0 0 0 0 0 0 0
## (80,85] (85,90] (90,95] (95,100]
## 0 0 0 12
Create Bins 2: decile of income?
- Suppose we want to put people into their respective income decile
inc.dec <- quantile(dat.inc$hhinc, prob = seq(0, 1, length = 11)) # income deciles
inc.dec
## 0% 10% 20% 30% 40% 50% 60% 70% 80%
## -993.1 132.6 193.3 250.4 314.5 392.3 482.0 576.4 703.8
## 90% 100%
## 924.9 38736.5
dat.inc$inc.cat <- cut(dat.inc$hhinc, inc.dec, labels = FALSE)
head(dat.inc[, c("hhinc", "inc.cat")]) # look at columns hhinc and inc.cat
## hhinc inc.cat
## 1 279.95 4
## 2 467.07 6
## 3 40.00 1
## 4 285.46 4
## 5 610.95 8
## 6 27.18 1
table(dat.inc$inc.cat)
##
## 1 2 3 4 5 6 7 8 9 10
## 678 678 679 678 679 678 678 679 678 679
Summaries by groups of Variables: library(plyr)
- Often we want summary statistics by groups of variables
tapply(), by() and aggregate() are base R possibilities- we’ll show
library(plyr) here
- click for the amazing tutorial of plyr by Hadley Wickham
- workhorse functions are
tranform(): transform a data.frame, (you get a df of same or bigger size)summarise(): summarises a data.frame (you get a df that is smaller)
summarise() and transform()
- use our FES data in
dat.inc.
- functions work on an entire data.frame
library(plyr)
# summarise an entire data.frame
summarise(dat.inc, mhhinc = mean(hhinc), vhhinc = sd(hhinc), mnumads = mean(numads))
## mhhinc vhhinc mnumads
## 1 499.2 649 1.797
# add 'income rank' to dat.inc. head(...,10) to see first 10 lines only
head(transform(dat.inc, inc.rank = rank(hhinc)), 10)
## hhref numads numadret numadern numhhkid kids0 kids1 kids2 kids34
## 1 1 1 1 0 0 0 0 0 0
## 2 2 2 0 2 1 0 0 0 0
## 3 3 1 0 0 0 0 0 0 0
## 4 4 2 1 1 0 0 0 0 0
## 5 5 3 0 3 1 0 0 0 0
## 6 6 1 0 1 0 0 0 0 0
## 7 7 2 0 2 1 1 0 0 0
## 8 8 2 0 2 0 0 0 0 0
## 9 9 1 0 0 0 0 0 0 0
## 10 10 4 0 3 0 0 0 0 0
## kids510 kids1116 kids1718 ncars nrooms age sex
## 1 0 0 0 1 6 66 female
## 2 0 1 0 0 4 38 male
## 3 0 0 0 0 3 25 male
## 4 0 0 0 0 3 80 female
## 5 0 1 0 2 6 40 male
## 6 0 0 0 1 4 50 female
## 7 0 0 0 1 5 29 male
## 8 0 0 0 1 4 26 female
## 9 0 0 0 0 3 27 male
## 10 0 0 0 2 5 41 female
## marstat ageced educ hours foodin foodout
## 1 Widowed 15 0 0 64.21 26.27
## 2 cohabiting 16 0 39 75.67 10.99
## 3 Single,nev marr inc wid,div,sep pre 91 0 6 0 11.86 6.67
## 4 Divorced 14 0 0 58.23 8.09
## 5 Married spouse in household 16 0 50 63.42 53.47
## 6 Divorced 15 0 5 19.73 5.32
## 7 Married spouse in household 21 0 30 36.97 7.65
## 8 cohabiting 17 0 38 11.64 22.36
## 9 Single,nev marr inc wid,div,sep pre 91 16 0 0 32.57 15.80
## 10 Married spouse in household 15 0 0 35.77 46.25
## alc nondur semidur durables wfoodin wfoodout walc ndex lndex
## 1 15.070 240.39 45.34 37.65 0.22474 0.09194 0.05274 285.73 5.655
## 2 26.075 257.56 10.50 13.99 0.28229 0.04100 0.09727 268.06 5.591
## 3 0.000 38.13 0.00 0.00 0.31104 0.17493 0.00000 38.13 3.641
## 4 23.610 142.49 226.82 86.35 0.15767 0.02191 0.06393 369.32 5.912
## 5 24.740 321.29 36.74 139.17 0.17713 0.14936 0.06910 358.02 5.881
## 6 19.905 79.15 0.00 0.00 0.24921 0.06721 0.25148 79.15 4.371
## 7 8.235 179.33 139.79 75.14 0.11586 0.02397 0.02581 319.12 5.766
## 8 21.995 182.79 2.62 13.96 0.06275 0.12059 0.11863 185.41 5.223
## 9 0.000 64.98 14.98 0.00 0.40733 0.19760 0.00000 79.96 4.382
## 10 37.025 387.78 68.24 15.45 0.07845 0.10142 0.08119 456.02 6.123
## year hhinc age.cat ced.cat inc.cat inc.rank
## 1 2005 279.95 (65,70] (10,15] 4 2361
## 2 2005 467.07 (35,40] (15,20] 6 3972
## 3 2005 40.00 (20,25] [0,5] 1 40
## 4 2005 285.46 (75,80] (10,15] 4 2419
## 5 2005 610.95 (35,40] (15,20] 8 4961
## 6 2005 27.18 (45,50] (10,15] 1 23
## 7 2005 639.37 (25,30] (20,25] 8 5109
## 8 2005 445.00 (25,30] (15,20] 6 3806
## 9 2005 91.92 (25,30] (15,20] 1 243
## 10 2005 772.30 (40,45] (10,15] 9 5679
transform() by groups: ddply()
- add income rank for each household for all categories of
marstat to dat
head(ddply(.data = dat.inc, .variables = "marstat", .fun = transform,
rank.marstat = rank(hhinc)), 10)
## hhref numads numadret numadern numhhkid kids0 kids1 kids2 kids34
## 1 5 3 0 3 1 0 0 0 0
## 2 7 2 0 2 1 1 0 0 0
## 3 10 4 0 3 0 0 0 0 0
## 4 12 2 0 1 0 0 0 0 0
## 5 13 2 0 2 2 0 0 0 0
## 6 18 2 2 0 0 0 0 0 0
## 7 20 3 0 1 0 0 0 0 0
## 8 21 2 2 0 0 0 0 0 0
## 9 24 2 0 1 0 0 0 0 0
## 10 25 2 0 1 1 0 1 0 0
## kids510 kids1116 kids1718 ncars nrooms age sex
## 1 0 1 0 2 6 40 male
## 2 0 0 0 1 5 29 male
## 3 0 0 0 2 5 41 female
## 4 0 0 0 1 4 50 male
## 5 0 2 0 1 5 40 male
## 6 0 0 0 1 5 73 male
## 7 0 0 0 1 6 59 male
## 8 0 0 0 0 6 73 male
## 9 0 0 0 0 5 56 male
## 10 0 0 0 1 4 22 female
## marstat ageced educ hours foodin foodout alc
## 1 Married spouse in household 16 0 50 63.42 53.47 24.740
## 2 Married spouse in household 21 0 30 36.97 7.65 8.235
## 3 Married spouse in household 15 0 0 35.77 46.25 37.025
## 4 Married spouse in household 20 0 0 6.53 9.72 0.000
## 5 Married spouse in household 16 0 40 16.14 32.59 56.785
## 6 Married spouse in household 14 0 0 35.47 20.24 2.495
## 7 Married spouse in household 15 0 0 107.10 13.27 99.505
## 8 Married spouse in household 15 0 0 47.58 11.15 25.190
## 9 Married spouse in household 16 0 37 44.26 7.35 19.630
## 10 Married spouse in household 16 0 0 16.50 18.53 0.000
## nondur semidur durables wfoodin wfoodout walc ndex lndex year
## 1 321.3 36.74 139.170 0.17713 0.14936 0.069102 358.0 5.881 2005
## 2 179.3 139.79 75.135 0.11586 0.02397 0.025805 319.1 5.766 2005
## 3 387.8 68.24 15.450 0.07845 0.10142 0.081193 456.0 6.123 2005
## 4 188.5 63.99 54.560 0.02586 0.03849 0.000000 252.5 5.532 2005
## 5 637.4 96.25 166.420 0.02200 0.04442 0.077396 733.7 6.598 2005
## 6 353.6 21.25 20.935 0.09462 0.05398 0.006656 374.9 5.927 2005
## 7 377.6 56.48 73.340 0.24673 0.03056 0.229237 434.1 6.073 2005
## 8 138.9 32.75 1.495 0.27723 0.06496 0.146759 171.6 5.145 2005
## 9 148.4 25.62 31.970 0.25431 0.04223 0.112777 174.1 5.159 2005
## 10 114.1 0.00 3.000 0.14470 0.16246 0.000000 114.1 4.737 2005
## hhinc age.cat ced.cat inc.cat rank.marstat
## 1 610.9 (35,40] (15,20] 8 1991
## 2 639.4 (25,30] (20,25] 8 2097
## 3 772.3 (40,45] (10,15] 9 2490
## 4 318.7 (45,50] (15,20] 5 710
## 5 737.4 (35,40] (15,20] 9 2407
## 6 283.2 (70,75] (10,15] 4 542
## 7 395.6 (55,60] (10,15] 6 1053
## 8 297.5 (70,75] (10,15] 4 621
## 9 334.8 (55,60] (15,20] 5 776
## 10 327.6 (20,25] (15,20] 5 747
- express everybody’s income as a percentage of the highest income in each category of
marstat
head(ddply(dat.inc, "marstat", transform, perc.of.max = hhinc/max(hhinc)),
10)
## hhref numads numadret numadern numhhkid kids0 kids1 kids2 kids34
## 1 5 3 0 3 1 0 0 0 0
## 2 7 2 0 2 1 1 0 0 0
## 3 10 4 0 3 0 0 0 0 0
## 4 12 2 0 1 0 0 0 0 0
## 5 13 2 0 2 2 0 0 0 0
## 6 18 2 2 0 0 0 0 0 0
## 7 20 3 0 1 0 0 0 0 0
## 8 21 2 2 0 0 0 0 0 0
## 9 24 2 0 1 0 0 0 0 0
## 10 25 2 0 1 1 0 1 0 0
## kids510 kids1116 kids1718 ncars nrooms age sex
## 1 0 1 0 2 6 40 male
## 2 0 0 0 1 5 29 male
## 3 0 0 0 2 5 41 female
## 4 0 0 0 1 4 50 male
## 5 0 2 0 1 5 40 male
## 6 0 0 0 1 5 73 male
## 7 0 0 0 1 6 59 male
## 8 0 0 0 0 6 73 male
## 9 0 0 0 0 5 56 male
## 10 0 0 0 1 4 22 female
## marstat ageced educ hours foodin foodout alc
## 1 Married spouse in household 16 0 50 63.42 53.47 24.740
## 2 Married spouse in household 21 0 30 36.97 7.65 8.235
## 3 Married spouse in household 15 0 0 35.77 46.25 37.025
## 4 Married spouse in household 20 0 0 6.53 9.72 0.000
## 5 Married spouse in household 16 0 40 16.14 32.59 56.785
## 6 Married spouse in household 14 0 0 35.47 20.24 2.495
## 7 Married spouse in household 15 0 0 107.10 13.27 99.505
## 8 Married spouse in household 15 0 0 47.58 11.15 25.190
## 9 Married spouse in household 16 0 37 44.26 7.35 19.630
## 10 Married spouse in household 16 0 0 16.50 18.53 0.000
## nondur semidur durables wfoodin wfoodout walc ndex lndex year
## 1 321.3 36.74 139.170 0.17713 0.14936 0.069102 358.0 5.881 2005
## 2 179.3 139.79 75.135 0.11586 0.02397 0.025805 319.1 5.766 2005
## 3 387.8 68.24 15.450 0.07845 0.10142 0.081193 456.0 6.123 2005
## 4 188.5 63.99 54.560 0.02586 0.03849 0.000000 252.5 5.532 2005
## 5 637.4 96.25 166.420 0.02200 0.04442 0.077396 733.7 6.598 2005
## 6 353.6 21.25 20.935 0.09462 0.05398 0.006656 374.9 5.927 2005
## 7 377.6 56.48 73.340 0.24673 0.03056 0.229237 434.1 6.073 2005
## 8 138.9 32.75 1.495 0.27723 0.06496 0.146759 171.6 5.145 2005
## 9 148.4 25.62 31.970 0.25431 0.04223 0.112777 174.1 5.159 2005
## 10 114.1 0.00 3.000 0.14470 0.16246 0.000000 114.1 4.737 2005
## hhinc age.cat ced.cat inc.cat perc.of.max
## 1 610.9 (35,40] (15,20] 8 0.015772
## 2 639.4 (25,30] (20,25] 8 0.016506
## 3 772.3 (40,45] (10,15] 9 0.019937
## 4 318.7 (45,50] (15,20] 5 0.008226
## 5 737.4 (35,40] (15,20] 9 0.019037
## 6 283.2 (70,75] (10,15] 4 0.007311
## 7 395.6 (55,60] (10,15] 6 0.010213
## 8 297.5 (70,75] (10,15] 4 0.007681
## 9 334.8 (55,60] (15,20] 5 0.008643
## 10 327.6 (20,25] (15,20] 5 0.008458
summarise() by groups
- median ratio of durable to nondurable expenditure by number of rooms
ddply(dat.inc, c("nrooms"), summarise, med.nondur.dur = median(nondur/durables,
na.rm = TRUE))
## nrooms med.nondur.dur
## 1 1 40.98
## 2 2 26.74
## 3 3 56.71
## 4 4 17.65
## 5 5 15.02
## 6 6 10.49
- by income decile
ddply(dat.inc, c("inc.cat"), summarise, med.nondur.dur = median(nondur/durables,
na.rm = T))
## inc.cat med.nondur.dur
## 1 1 55.923
## 2 2 46.671
## 3 3 20.446
## 4 4 16.653
## 5 5 17.453
## 6 6 11.652
## 7 7 10.394
## 8 8 7.555
## 9 9 7.727
## 10 10 6.765
## 11 NA 31.173
- by both
ddply(dat.inc, c("nrooms", "inc.cat"), summarise, med.nondur.dur = median(nondur/durables))
## nrooms inc.cat med.nondur.dur
## 1 1 1 29.514
## 2 1 2 3.441
## 3 1 3 Inf
## 4 1 4 Inf
## 5 2 1 55.923
## 6 2 2 264.305
## 7 2 3 70.520
## 8 2 4 14.303
## 9 2 5 95.747
## 10 2 6 26.736
## 11 2 7 5.952
## 12 2 8 4.920
## 13 2 9 13.812
## 14 2 10 Inf
## 15 3 1 NA
## 16 3 2 68.800
## 17 3 3 154.473
## 18 3 4 22.531
## 19 3 5 66.409
## 20 3 6 34.023
## 21 3 7 24.682
## 22 3 8 29.883
## 23 3 9 14.360
## 24 3 10 Inf
## 25 4 1 46.028
## 26 4 2 56.398
## 27 4 3 16.572
## 28 4 4 21.661
## 29 4 5 19.852
## 30 4 6 12.512
## 31 4 7 9.620
## 32 4 8 7.562
## 33 4 9 6.118
## 34 4 10 7.868
## 35 5 1 54.167
## 36 5 2 57.002
## 37 5 3 22.446
## 38 5 4 13.884
## 39 5 5 13.223
## 40 5 6 9.310
## 41 5 7 10.243
## 42 5 8 6.880
## 43 5 9 9.338
## 44 5 10 8.861
## 45 6 1 48.133
## 46 6 2 34.481
## 47 6 3 11.674
## 48 6 4 17.718
## 49 6 5 15.791
## 50 6 6 11.948
## 51 6 7 10.505
## 52 6 8 7.740
## 53 6 9 7.374
## 54 6 10 6.549
## 55 6 NA 31.173
more ddply{plyr}
- Split data frame, apply function, and return results in a data frame.
library(plyr)
ddply(.data = dat.inc, .variables = "sex", .fun = summarise, inc = mean(hhinc)) # mean income by sex?
## sex inc
## 1 male 552.9
## 2 female 424.0
ddply(dat.inc, c("sex", "numads"), summarise, alc = mean(alc), foodin = mean(foodin),
foodout = mean(foodout))
## sex numads alc foodin foodout
## 1 male 1 12.335 21.64 12.512
## 2 male 2 16.085 53.34 24.060
## 3 male 3 24.062 68.01 35.314
## 4 male 4 36.492 74.62 50.590
## 5 male 5 30.686 109.28 54.595
## 6 male 6 54.084 133.02 81.595
## 7 male 7 0.000 158.16 5.625
## 8 male 8 0.000 82.87 35.075
## 9 male 9 156.450 95.94 25.695
## 10 female 1 4.710 28.72 9.080
## 11 female 2 16.455 51.37 24.505
## 12 female 3 23.975 60.93 35.894
## 13 female 4 30.336 81.28 42.586
## 14 female 5 39.619 81.89 49.511
## 15 female 6 6.657 85.10 54.789
ddply{plyr} 2
summarise can be many things, not just mean()- you can supply any function you want
- check out Hadley’s tutorial.
ddply is very powerful.
ddply(dat.inc, "sex", summarise, age.range = median(age), car.rooms = max(ncars/nrooms))
## sex age.range car.rooms
## 1 male 52 1.4
## 2 female 50 1.0
ddply(subset(dat.inc, numhhkid > 1), "ncars", function(x) data.frame(dur.inc = mean(x$dur/x$hhinc),
nondur.inc = mean(x$nondur/x$hhinc))) # instead of summarise(), supply an 'anonymous' function
## ncars dur.inc nondur.inc
## 1 0 0.105007 0.6959
## 2 1 0.114872 0.6514
## 3 2 0.131991 0.6045
## 4 3 0.144138 0.6760
## 5 4 0.175777 1.5216
## 6 5 0.148054 0.5776
## 7 7 0.005687 0.6105
Alternatives to ddply
- very powerful and elegent package:
data.table
- a
data.table is a data.frame on speed
- excellent documentation and explainers (vignette) at CRAN
- very fast
- maybe easier to understand than ddply
- read the 10 minutes intro on the CRAN link above
summary by group with data.table
- a data.table has two indices,
i and j: DT[i,j]
i is SQL where and j is an expression
library(data.table)
dat.tab <- data.table(dat.inc)
tables()
## NAME NROW MB
## [1,] dat.tab 6,785 2
## COLS
## [1,] hhref,numads,numadret,numadern,numhhkid,kids0,kids1,kids2,kids34,kids510,kids111
## KEY
## [1,]
## Total: 2MB
dat.tab[, list(median.age = as.numeric(median(age)), cars.over.rooms = median(ncars/nrooms)),
by = sex]
## sex median.age cars.over.rooms
## 1: female 50 0.1667
## 2: male 52 0.2000
# adding a column 'by reference' with :=
dat.tab[, `:=`(rel.inc, hhinc/median(hhinc))] # I typed 'rel.inc := hhinc/median(hhinc)' here.
## hhref numads numadret numadern numhhkid kids0 kids1 kids2 kids34
## 1: 1 1 1 0 0 0 0 0 0
## 2: 2 2 0 2 1 0 0 0 0
## 3: 3 1 0 0 0 0 0 0 0
## 4: 4 2 1 1 0 0 0 0 0
## 5: 5 3 0 3 1 0 0 0 0
## ---
## 6781: 6781 4 0 3 2 0 0 0 0
## 6782: 6782 2 0 1 2 0 0 0 0
## 6783: 6783 1 0 0 2 0 0 0 0
## 6784: 6784 1 1 0 0 0 0 0 0
## 6785: 6785 1 0 0 1 0 0 1 0
## kids510 kids1116 kids1718 ncars nrooms age sex
## 1: 0 0 0 1 6 66 female
## 2: 0 1 0 0 4 38 male
## 3: 0 0 0 0 3 25 male
## 4: 0 0 0 0 3 80 female
## 5: 0 1 0 2 6 40 male
## ---
## 6781: 0 1 1 3 6 42 male
## 6782: 0 1 1 1 6 49 male
## 6783: 0 2 0 1 5 33 female
## 6784: 0 0 0 0 5 76 female
## 6785: 0 0 0 0 5 24 female
## marstat ageced educ hours foodin
## 1: Widowed 15 0 0 64.21
## 2: cohabiting 16 0 39 75.67
## 3: Single,nev marr inc wid,div,sep pre 91 0 6 0 11.86
## 4: Divorced 14 0 0 58.23
## 5: Married spouse in household 16 0 50 63.42
## ---
## 6781: Married spouse in household 16 0 0 122.00
## 6782: Married spouse in household 16 0 0 22.48
## 6783: Divorced 18 0 0 18.45
## 6784: Widowed 15 0 0 35.31
## 6785: Single,nev marr inc wid,div,sep pre 91 16 0 0 0.00
## foodout alc nondur semidur durables wfoodin wfoodout walc
## 1: 26.27 15.07 240.39 45.34 37.650 0.22474 0.09194 0.05274
## 2: 10.99 26.07 257.56 10.50 13.995 0.28229 0.04100 0.09727
## 3: 6.67 0.00 38.13 0.00 0.000 0.31104 0.17493 0.00000
## 4: 8.09 23.61 142.49 226.82 86.355 0.15767 0.02191 0.06393
## 5: 53.47 24.74 321.29 36.74 139.170 0.17713 0.14936 0.06910
## ---
## 6781: 50.25 120.50 641.52 64.52 98.320 0.17280 0.07117 0.17067
## 6782: 88.90 31.35 393.19 71.12 9.620 0.04840 0.19147 0.06751
## 6783: 5.90 0.00 206.65 67.00 6.995 0.06744 0.02156 0.00000
## 6784: 0.00 0.00 95.53 0.00 1.255 0.36964 0.00000 0.00000
## 6785: 0.00 0.00 97.00 0.00 165.000 0.00000 0.00000 0.00000
## ndex lndex year hhinc age.cat ced.cat inc.cat rel.inc
## 1: 285.73 5.655 2005 280.0 (65,70] (10,15] 4 0.7136
## 2: 268.06 5.591 2005 467.1 (35,40] (15,20] 6 1.1905
## 3: 38.13 3.641 2005 40.0 (20,25] [0,5] 1 0.1020
## 4: 369.32 5.912 2005 285.5 (75,80] (10,15] 4 0.7276
## 5: 358.02 5.881 2005 610.9 (35,40] (15,20] 8 1.5572
## ---
## 6781: 706.04 6.560 2005 1184.2 (40,45] (15,20] 10 3.0185
## 6782: 464.31 6.141 2005 388.2 (45,50] (15,20] 5 0.9896
## 6783: 273.65 5.612 2005 248.6 (30,35] (15,20] 3 0.6337
## 6784: 95.53 4.559 2005 113.5 (75,80] (10,15] 1 0.2893
## 6785: 97.00 4.575 2005 120.0 (20,25] (15,20] 1 0.3059
dat.tab[, `:=`(rel.inc.sex, hhinc/median(hhinc)), by = sex]
## hhref numads numadret numadern numhhkid kids0 kids1 kids2 kids34
## 1: 1 1 1 0 0 0 0 0 0
## 2: 2 2 0 2 1 0 0 0 0
## 3: 3 1 0 0 0 0 0 0 0
## 4: 4 2 1 1 0 0 0 0 0
## 5: 5 3 0 3 1 0 0 0 0
## ---
## 6781: 6781 4 0 3 2 0 0 0 0
## 6782: 6782 2 0 1 2 0 0 0 0
## 6783: 6783 1 0 0 2 0 0 0 0
## 6784: 6784 1 1 0 0 0 0 0 0
## 6785: 6785 1 0 0 1 0 0 1 0
## kids510 kids1116 kids1718 ncars nrooms age sex
## 1: 0 0 0 1 6 66 female
## 2: 0 1 0 0 4 38 male
## 3: 0 0 0 0 3 25 male
## 4: 0 0 0 0 3 80 female
## 5: 0 1 0 2 6 40 male
## ---
## 6781: 0 1 1 3 6 42 male
## 6782: 0 1 1 1 6 49 male
## 6783: 0 2 0 1 5 33 female
## 6784: 0 0 0 0 5 76 female
## 6785: 0 0 0 0 5 24 female
## marstat ageced educ hours foodin
## 1: Widowed 15 0 0 64.21
## 2: cohabiting 16 0 39 75.67
## 3: Single,nev marr inc wid,div,sep pre 91 0 6 0 11.86
## 4: Divorced 14 0 0 58.23
## 5: Married spouse in household 16 0 50 63.42
## ---
## 6781: Married spouse in household 16 0 0 122.00
## 6782: Married spouse in household 16 0 0 22.48
## 6783: Divorced 18 0 0 18.45
## 6784: Widowed 15 0 0 35.31
## 6785: Single,nev marr inc wid,div,sep pre 91 16 0 0 0.00
## foodout alc nondur semidur durables wfoodin wfoodout walc
## 1: 26.27 15.07 240.39 45.34 37.650 0.22474 0.09194 0.05274
## 2: 10.99 26.07 257.56 10.50 13.995 0.28229 0.04100 0.09727
## 3: 6.67 0.00 38.13 0.00 0.000 0.31104 0.17493 0.00000
## 4: 8.09 23.61 142.49 226.82 86.355 0.15767 0.02191 0.06393
## 5: 53.47 24.74 321.29 36.74 139.170 0.17713 0.14936 0.06910
## ---
## 6781: 50.25 120.50 641.52 64.52 98.320 0.17280 0.07117 0.17067
## 6782: 88.90 31.35 393.19 71.12 9.620 0.04840 0.19147 0.06751
## 6783: 5.90 0.00 206.65 67.00 6.995 0.06744 0.02156 0.00000
## 6784: 0.00 0.00 95.53 0.00 1.255 0.36964 0.00000 0.00000
## 6785: 0.00 0.00 97.00 0.00 165.000 0.00000 0.00000 0.00000
## ndex lndex year hhinc age.cat ced.cat inc.cat rel.inc rel.inc.sex
## 1: 285.73 5.655 2005 280.0 (65,70] (10,15] 4 0.7136 0.92498
## 2: 268.06 5.591 2005 467.1 (35,40] (15,20] 6 1.1905 1.01540
## 3: 38.13 3.641 2005 40.0 (20,25] [0,5] 1 0.1020 0.08696
## 4: 369.32 5.912 2005 285.5 (75,80] (10,15] 4 0.7276 0.94320
## 5: 358.02 5.881 2005 610.9 (35,40] (15,20] 8 1.5572 1.32818
## ---
## 6781: 706.04 6.560 2005 1184.2 (40,45] (15,20] 10 3.0185 2.57446
## 6782: 464.31 6.141 2005 388.2 (45,50] (15,20] 5 0.9896 0.84404
## 6783: 273.65 5.612 2005 248.6 (30,35] (15,20] 3 0.6337 0.82140
## 6784: 95.53 4.559 2005 113.5 (75,80] (10,15] 1 0.2893 0.37499
## 6785: 97.00 4.575 2005 120.0 (20,25] (15,20] 1 0.3059 0.39649
dat.tab[, `:=`(rel.inc.sex.nrooms, hhinc/median(hhinc)), by = list(sex,
nrooms)]
## hhref numads numadret numadern numhhkid kids0 kids1 kids2 kids34
## 1: 1 1 1 0 0 0 0 0 0
## 2: 2 2 0 2 1 0 0 0 0
## 3: 3 1 0 0 0 0 0 0 0
## 4: 4 2 1 1 0 0 0 0 0
## 5: 5 3 0 3 1 0 0 0 0
## ---
## 6781: 6781 4 0 3 2 0 0 0 0
## 6782: 6782 2 0 1 2 0 0 0 0
## 6783: 6783 1 0 0 2 0 0 0 0
## 6784: 6784 1 1 0 0 0 0 0 0
## 6785: 6785 1 0 0 1 0 0 1 0
## kids510 kids1116 kids1718 ncars nrooms age sex
## 1: 0 0 0 1 6 66 female
## 2: 0 1 0 0 4 38 male
## 3: 0 0 0 0 3 25 male
## 4: 0 0 0 0 3 80 female
## 5: 0 1 0 2 6 40 male
## ---
## 6781: 0 1 1 3 6 42 male
## 6782: 0 1 1 1 6 49 male
## 6783: 0 2 0 1 5 33 female
## 6784: 0 0 0 0 5 76 female
## 6785: 0 0 0 0 5 24 female
## marstat ageced educ hours foodin
## 1: Widowed 15 0 0 64.21
## 2: cohabiting 16 0 39 75.67
## 3: Single,nev marr inc wid,div,sep pre 91 0 6 0 11.86
## 4: Divorced 14 0 0 58.23
## 5: Married spouse in household 16 0 50 63.42
## ---
## 6781: Married spouse in household 16 0 0 122.00
## 6782: Married spouse in household 16 0 0 22.48
## 6783: Divorced 18 0 0 18.45
## 6784: Widowed 15 0 0 35.31
## 6785: Single,nev marr inc wid,div,sep pre 91 16 0 0 0.00
## foodout alc nondur semidur durables wfoodin wfoodout walc
## 1: 26.27 15.07 240.39 45.34 37.650 0.22474 0.09194 0.05274
## 2: 10.99 26.07 257.56 10.50 13.995 0.28229 0.04100 0.09727
## 3: 6.67 0.00 38.13 0.00 0.000 0.31104 0.17493 0.00000
## 4: 8.09 23.61 142.49 226.82 86.355 0.15767 0.02191 0.06393
## 5: 53.47 24.74 321.29 36.74 139.170 0.17713 0.14936 0.06910
## ---
## 6781: 50.25 120.50 641.52 64.52 98.320 0.17280 0.07117 0.17067
## 6782: 88.90 31.35 393.19 71.12 9.620 0.04840 0.19147 0.06751
## 6783: 5.90 0.00 206.65 67.00 6.995 0.06744 0.02156 0.00000
## 6784: 0.00 0.00 95.53 0.00 1.255 0.36964 0.00000 0.00000
## 6785: 0.00 0.00 97.00 0.00 165.000 0.00000 0.00000 0.00000
## ndex lndex year hhinc age.cat ced.cat inc.cat rel.inc rel.inc.sex
## 1: 285.73 5.655 2005 280.0 (65,70] (10,15] 4 0.7136 0.92498
## 2: 268.06 5.591 2005 467.1 (35,40] (15,20] 6 1.1905 1.01540
## 3: 38.13 3.641 2005 40.0 (20,25] [0,5] 1 0.1020 0.08696
## 4: 369.32 5.912 2005 285.5 (75,80] (10,15] 4 0.7276 0.94320
## 5: 358.02 5.881 2005 610.9 (35,40] (15,20] 8 1.5572 1.32818
## ---
## 6781: 706.04 6.560 2005 1184.2 (40,45] (15,20] 10 3.0185 2.57446
## 6782: 464.31 6.141 2005 388.2 (45,50] (15,20] 5 0.9896 0.84404
## 6783: 273.65 5.612 2005 248.6 (30,35] (15,20] 3 0.6337 0.82140
## 6784: 95.53 4.559 2005 113.5 (75,80] (10,15] 1 0.2893 0.37499
## 6785: 97.00 4.575 2005 120.0 (20,25] (15,20] 1 0.3059 0.39649
## rel.inc.sex.nrooms
## 1: 0.6235
## 2: 1.4289
## 3: 0.1760
## 4: 1.6725
## 5: 1.0829
## ---
## 6781: 2.0990
## 6782: 0.6882
## 6783: 0.9589
## 6784: 0.4378
## 6785: 0.4629
data.tables and keys
- very large speed gain by avoiding vector scan and using binary search instead
- you set a key on a table, like a database
setkey(dat.tab, numhhkid)
dat.tab[numhhkid == 1]
## hhref numads numadret numadern numhhkid kids0 kids1 kids2 kids34
## 1: 2 2 0 2 1 0 0 0 0
## 2: 5 3 0 3 1 0 0 0 0
## 3: 7 2 0 2 1 1 0 0 0
## 4: 17 2 0 0 1 0 0 0 1
## 5: 25 2 0 1 1 0 1 0 0
## ---
## 898: 6751 2 0 2 1 0 0 0 0
## 899: 6765 2 0 1 1 0 0 0 1
## 900: 6776 2 0 1 1 0 0 0 1
## 901: 6777 2 0 2 1 0 0 0 0
## 902: 6785 1 0 0 1 0 0 1 0
## kids510 kids1116 kids1718 ncars nrooms age sex
## 1: 0 1 0 0 4 38 male
## 2: 0 1 0 2 6 40 male
## 3: 0 0 0 1 5 29 male
## 4: 0 0 0 0 5 22 female
## 5: 0 0 0 1 4 22 female
## ---
## 898: 1 0 0 2 6 57 male
## 899: 0 0 0 1 6 33 male
## 900: 0 0 0 1 6 25 male
## 901: 0 1 0 2 6 47 male
## 902: 0 0 0 0 5 24 female
## marstat ageced educ hours foodin
## 1: cohabiting 16 0 39 75.67
## 2: Married spouse in household 16 0 50 63.42
## 3: Married spouse in household 21 0 30 36.97
## 4: cohabiting 16 0 0 10.33
## 5: Married spouse in household 16 0 0 16.50
## ---
## 898: Married spouse in household 21 0 40 148.51
## 899: cohabiting 19 0 0 46.60
## 900: Married spouse in household 18 0 38 35.83
## 901: Married spouse in household 21 0 37 100.82
## 902: Single,nev marr inc wid,div,sep pre 91 16 0 0 0.00
## foodout alc nondur semidur durables wfoodin wfoodout walc
## 1: 10.99 26.075 257.56 10.50 13.99 0.2823 0.04100 0.097274
## 2: 53.47 24.740 321.29 36.74 139.17 0.1771 0.14936 0.069102
## 3: 7.65 8.235 179.33 139.79 75.14 0.1159 0.02397 0.025805
## 4: 5.37 0.000 48.88 8.00 0.00 0.1816 0.09440 0.000000
## 5: 18.53 0.000 114.06 0.00 3.00 0.1447 0.16246 0.000000
## ---
## 898: 47.80 2.590 633.84 56.62 0.58 0.2151 0.06923 0.003751
## 899: 33.55 36.765 288.63 77.24 107.65 0.1274 0.09170 0.100487
## 900: 72.55 29.500 248.35 89.00 9.69 0.1062 0.21506 0.087448
## 901: 56.49 0.000 735.15 136.02 70.88 0.1157 0.06485 0.000000
## 902: 0.00 0.000 97.00 0.00 165.00 0.0000 0.00000 0.000000
## ndex lndex year hhinc age.cat ced.cat inc.cat rel.inc rel.inc.sex
## 1: 268.06 5.591 2005 467.1 (35,40] (15,20] 6 1.1905 1.0154
## 2: 358.02 5.881 2005 610.9 (35,40] (15,20] 8 1.5572 1.3282
## 3: 319.12 5.766 2005 639.4 (25,30] (20,25] 8 1.6297 1.3900
## 4: 56.88 4.041 2005 182.2 (20,25] (15,20] 2 0.4645 0.6021
## 5: 114.06 4.737 2005 327.6 (20,25] (15,20] 5 0.8351 1.0826
## ---
## 898: 690.46 6.537 2005 1192.1 (55,60] (20,25] 10 3.0385 2.5915
## 899: 365.87 5.902 2005 405.8 (30,35] (15,20] 6 1.0344 0.8823
## 900: 337.35 5.821 2005 321.6 (20,25] (15,20] 5 0.8198 0.6992
## 901: 871.17 6.770 2005 903.6 (45,50] (20,25] 9 2.3032 1.9644
## 902: 97.00 4.575 2005 120.0 (20,25] (15,20] 1 0.3059 0.3965
## rel.inc.sex.nrooms
## 1: 1.4289
## 2: 1.0829
## 3: 1.5904
## 4: 0.7028
## 5: 1.4260
## ---
## 898: 2.1129
## 899: 0.7193
## 900: 0.5701
## 901: 1.6016
## 902: 0.4629
dat.tab[J(1)]
## numhhkid hhref numads numadret numadern kids0 kids1 kids2 kids34
## 1: 1 2 2 0 2 0 0 0 0
## 2: 1 5 3 0 3 0 0 0 0
## 3: 1 7 2 0 2 1 0 0 0
## 4: 1 17 2 0 0 0 0 0 1
## 5: 1 25 2 0 1 0 1 0 0
## ---
## 898: 1 6751 2 0 2 0 0 0 0
## 899: 1 6765 2 0 1 0 0 0 1
## 900: 1 6776 2 0 1 0 0 0 1
## 901: 1 6777 2 0 2 0 0 0 0
## 902: 1 6785 1 0 0 0 0 1 0
## kids510 kids1116 kids1718 ncars nrooms age sex
## 1: 0 1 0 0 4 38 male
## 2: 0 1 0 2 6 40 male
## 3: 0 0 0 1 5 29 male
## 4: 0 0 0 0 5 22 female
## 5: 0 0 0 1 4 22 female
## ---
## 898: 1 0 0 2 6 57 male
## 899: 0 0 0 1 6 33 male
## 900: 0 0 0 1 6 25 male
## 901: 0 1 0 2 6 47 male
## 902: 0 0 0 0 5 24 female
## marstat ageced educ hours foodin
## 1: cohabiting 16 0 39 75.67
## 2: Married spouse in household 16 0 50 63.42
## 3: Married spouse in household 21 0 30 36.97
## 4: cohabiting 16 0 0 10.33
## 5: Married spouse in household 16 0 0 16.50
## ---
## 898: Married spouse in household 21 0 40 148.51
## 899: cohabiting 19 0 0 46.60
## 900: Married spouse in household 18 0 38 35.83
## 901: Married spouse in household 21 0 37 100.82
## 902: Single,nev marr inc wid,div,sep pre 91 16 0 0 0.00
## foodout alc nondur semidur durables wfoodin wfoodout walc
## 1: 10.99 26.075 257.56 10.50 13.99 0.2823 0.04100 0.097274
## 2: 53.47 24.740 321.29 36.74 139.17 0.1771 0.14936 0.069102
## 3: 7.65 8.235 179.33 139.79 75.14 0.1159 0.02397 0.025805
## 4: 5.37 0.000 48.88 8.00 0.00 0.1816 0.09440 0.000000
## 5: 18.53 0.000 114.06 0.00 3.00 0.1447 0.16246 0.000000
## ---
## 898: 47.80 2.590 633.84 56.62 0.58 0.2151 0.06923 0.003751
## 899: 33.55 36.765 288.63 77.24 107.65 0.1274 0.09170 0.100487
## 900: 72.55 29.500 248.35 89.00 9.69 0.1062 0.21506 0.087448
## 901: 56.49 0.000 735.15 136.02 70.88 0.1157 0.06485 0.000000
## 902: 0.00 0.000 97.00 0.00 165.00 0.0000 0.00000 0.000000
## ndex lndex year hhinc age.cat ced.cat inc.cat rel.inc rel.inc.sex
## 1: 268.06 5.591 2005 467.1 (35,40] (15,20] 6 1.1905 1.0154
## 2: 358.02 5.881 2005 610.9 (35,40] (15,20] 8 1.5572 1.3282
## 3: 319.12 5.766 2005 639.4 (25,30] (20,25] 8 1.6297 1.3900
## 4: 56.88 4.041 2005 182.2 (20,25] (15,20] 2 0.4645 0.6021
## 5: 114.06 4.737 2005 327.6 (20,25] (15,20] 5 0.8351 1.0826
## ---
## 898: 690.46 6.537 2005 1192.1 (55,60] (20,25] 10 3.0385 2.5915
## 899: 365.87 5.902 2005 405.8 (30,35] (15,20] 6 1.0344 0.8823
## 900: 337.35 5.821 2005 321.6 (20,25] (15,20] 5 0.8198 0.6992
## 901: 871.17 6.770 2005 903.6 (45,50] (20,25] 9 2.3032 1.9644
## 902: 97.00 4.575 2005 120.0 (20,25] (15,20] 1 0.3059 0.3965
## rel.inc.sex.nrooms
## 1: 1.4289
## 2: 1.0829
## 3: 1.5904
## 4: 0.7028
## 5: 1.4260
## ---
## 898: 2.1129
## 899: 0.7193
## 900: 0.5701
## 901: 1.6016
## 902: 0.4629
dat.tab[J(2), `:=`(twokid.foodin, log(foodin)/log(foodout))]
## hhref numads numadret numadern numhhkid kids0 kids1 kids2 kids34
## 1: 1 1 1 0 0 0 0 0 0
## 2: 3 1 0 0 0 0 0 0 0
## 3: 4 2 1 1 0 0 0 0 0
## 4: 6 1 0 1 0 0 0 0 0
## 5: 8 2 0 2 0 0 0 0 0
## ---
## 6781: 3230 3 0 2 7 0 1 0 1
## 6782: 3466 6 0 0 7 0 0 1 5
## 6783: 4824 4 0 3 7 0 0 0 2
## 6784: 6629 2 0 1 7 0 0 0 1
## 6785: 4787 1 0 1 8 1 0 2 0
## kids510 kids1116 kids1718 ncars nrooms age sex
## 1: 0 0 0 1 6 66 female
## 2: 0 0 0 0 3 25 male
## 3: 0 0 0 0 3 80 female
## 4: 0 0 0 1 4 50 female
## 5: 0 0 0 1 4 26 female
## ---
## 6781: 2 3 0 0 5 43 male
## 6782: 0 0 1 1 6 43 female
## 6783: 3 2 0 3 6 41 female
## 6784: 4 2 0 1 6 46 male
## 6785: 2 3 0 0 6 36 female
## marstat ageced educ hours foodin
## 1: Widowed 15 0 0 64.21
## 2: Single,nev marr inc wid,div,sep pre 91 0 6 0 11.86
## 3: Divorced 14 0 0 58.23
## 4: Divorced 15 0 5 19.73
## 5: cohabiting 17 0 38 11.64
## ---
## 6781: Married spouse in household 20 0 36 23.97
## 6782: Separated 97 0 0 153.38
## 6783: Married spouse in household 16 0 0 85.31
## 6784: Married spouse in household 16 0 44 95.18
## 6785: Single,nev marr inc wid,div,sep pre 91 16 0 20 92.93
## foodout alc nondur semidur durables wfoodin wfoodout walc
## 1: 26.27 15.07 240.39 45.34 37.65 0.22474 0.09194 0.05274
## 2: 6.67 0.00 38.13 0.00 0.00 0.31104 0.17493 0.00000
## 3: 8.09 23.61 142.49 226.82 86.35 0.15767 0.02191 0.06393
## 4: 5.32 19.91 79.15 0.00 0.00 0.24921 0.06721 0.25148
## 5: 22.36 22.00 182.79 2.62 13.96 0.06275 0.12059 0.11863
## ---
## 6781: 11.17 0.00 118.97 16.49 3.08 0.17690 0.08249 0.00000
## 6782: 76.65 0.00 479.57 123.40 276.12 0.25438 0.12712 0.00000
## 6783: 30.32 8.72 498.84 32.83 105.32 0.16045 0.05704 0.01640
## 6784: 10.77 13.70 328.66 15.83 0.00 0.27631 0.03126 0.03977
## 6785: 46.83 4.49 317.51 40.85 51.16 0.25931 0.13068 0.01253
## ndex lndex year hhinc age.cat ced.cat inc.cat rel.inc
## 1: 285.73 5.655 2005 279.95 (65,70] (10,15] 4 0.71356
## 2: 38.13 3.641 2005 40.00 (20,25] [0,5] 1 0.10195
## 3: 369.32 5.912 2005 285.46 (75,80] (10,15] 4 0.72761
## 4: 79.15 4.371 2005 27.18 (45,50] (10,15] 1 0.06928
## 5: 185.41 5.223 2005 445.00 (25,30] (15,20] 6 1.13425
## ---
## 6781: 135.47 4.909 2005 482.50 (40,45] (15,20] 7 1.22983
## 6782: 602.97 6.402 2005 611.20 (40,45] (95,100] 8 1.55787
## 6783: 531.67 6.276 2005 1576.87 (40,45] (15,20] 10 4.01924
## 6784: 344.49 5.842 2005 541.65 (45,50] (15,20] 7 1.38060
## 6785: 358.35 5.882 2005 518.42 (35,40] (15,20] 7 1.32139
## rel.inc.sex rel.inc.sex.nrooms twokid.foodin
## 1: 0.92498 0.6235 NA
## 2: 0.08696 0.1760 NA
## 3: 0.94320 1.6725 NA
## 4: 0.08981 0.1183 NA
## 5: 1.47033 1.9368 NA
## ---
## 6781: 1.04893 1.2002 NA
## 6782: 2.01947 1.3612 NA
## 6783: 5.21014 3.5119 NA
## 6784: 1.17752 0.9601 NA
## 6785: 1.71291 1.1546 NA
Graphics and R
- production of high-quality graphics is another strength of R
- base R is a good start. Many class-specific plotting methods.
- more functionality from other packages:
library(ggplot2)library(lattice)- one could easily fill 2 hours talking about ggplot alone. Barely scratching the surface here.
Base R plots
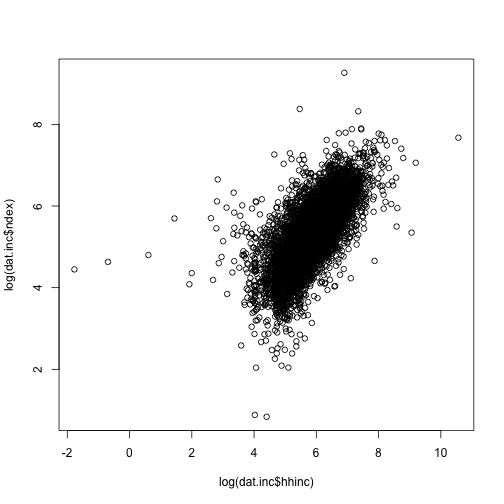
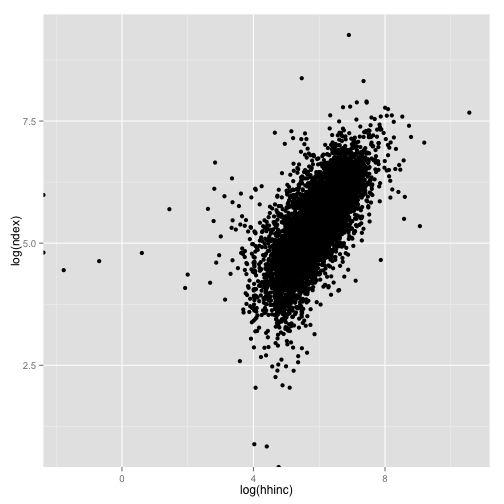
plot(y = log(dat.inc$ndex), x = log(dat.inc$hhinc)) # many options. see ?plot and ?par
## Warning: NaNs produced
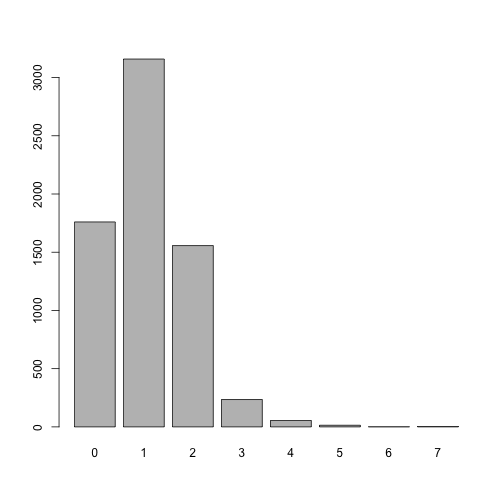
plot(factor(dat.inc$ncars)) # plot depends on data types
Plots with ggplot
- ggplots are based on the Grammar of Graphics
- click here for help, examples and tutorials at http://ggplot2.org/
- a plot is defined by a set of data that is mapped onto a picture - in many complicated ways, if you so desire
- the mappings are called
aes() thetics. For example aes(x=var1,y=var2)
- the way they appear in the picture are called
geom etrics
- there is a very long list of
geoms. Refer to documentation.
- let’s plot log non-durable expenditure against log household income in a scatter plot
library(ggplot2)
p <- ggplot(data = dat.inc, aes(x = log(hhinc), y = log(ndex))) # not a plot yet: misses a geom
p + geom_point() # now that's a plot
## Warning: NaNs produced
## Warning: NaNs produced
## Warning: Removed 4 rows containing missing values (geom_point).
ggplot 2
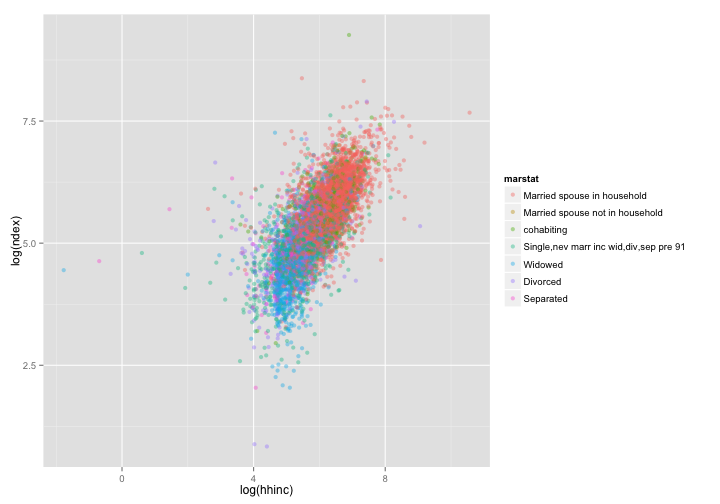
dat <- subset(dat.inc, hhinc > 0 & ndex > 0) # get rid of some negative values
# add another layer: color points by marstat, and decrease opacity to 0.4
p <- ggplot(data = dat, aes(x = log(hhinc), y = log(ndex))) # not a plot yet: misses a geom
p + geom_point(aes(color = marstat), alpha = 0.4)
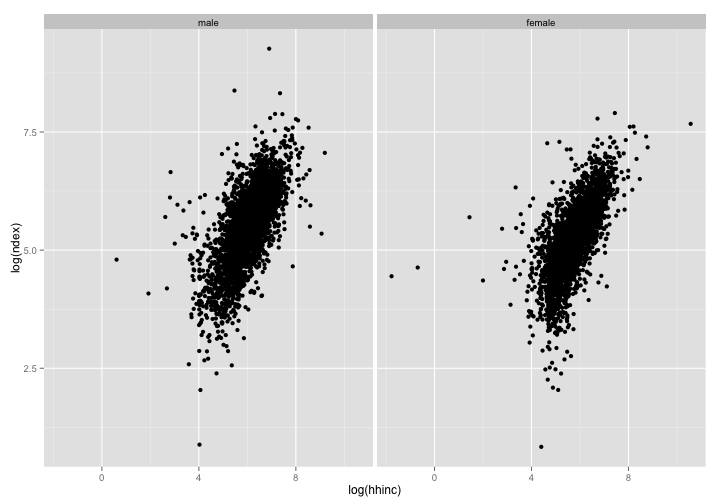
p + geom_point() + facet_wrap(~sex) # split plot by factor sex
More ggplots
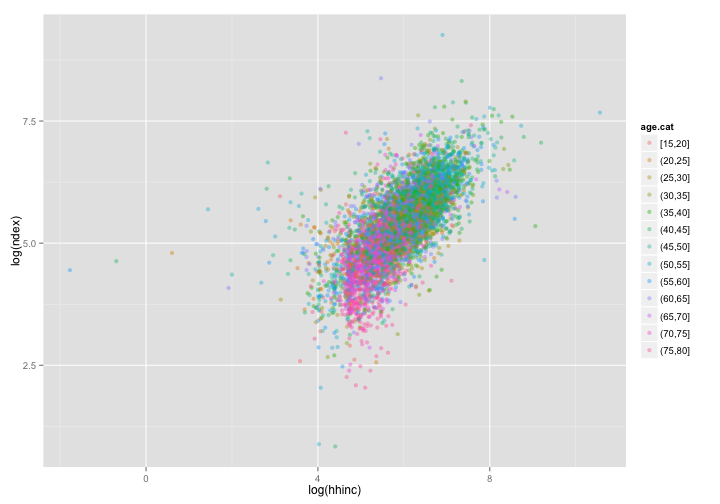
ggplot(data = dat, aes(x = log(hhinc), y = log(ndex))) + geom_point(aes(color = age.cat),
alpha = 0.4)
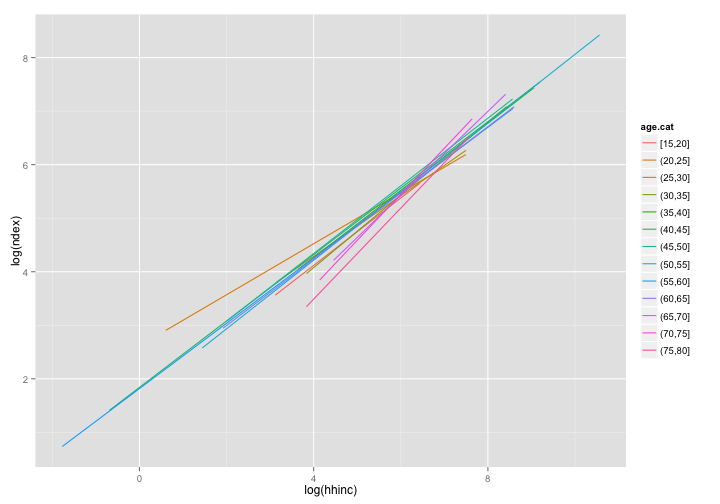
ggplot(data = dat, aes(x = log(hhinc), y = log(ndex))) + geom_smooth(aes(color = age.cat),
method = "lm", se = FALSE)
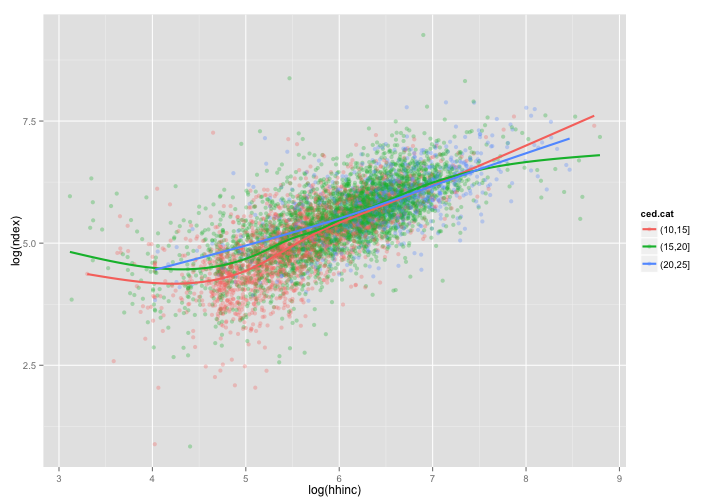
sdat <- subset(dat, ced.cat %in% c("(10,15]", "(15,20]", "(20,25]") &
log(hhinc) > 3 & log(hhinc) < 9) # select a subset of ced
ggplot(data = sdat, aes(x = log(hhinc), y = log(ndex))) + geom_point(aes(color = ced.cat),
alpha = 0.3) + geom_smooth(aes(color = ced.cat), se = FALSE, size = 1)
## geom_smooth: method="auto" and size of largest group is >=1000, so using
## gam with formula: y ~ s(x, bs = "cs"). Use 'method = x' to change the
## smoothing method.
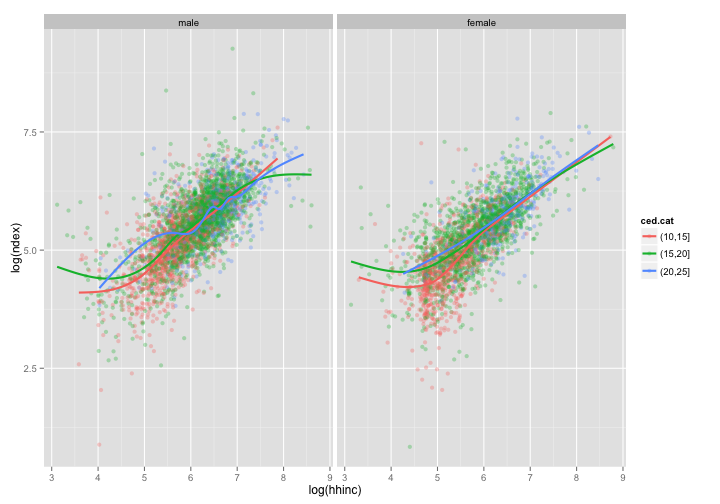
# split by sex?
ggplot(data = sdat, aes(x = log(hhinc), y = log(ndex))) + geom_point(aes(color = ced.cat),
alpha = 0.3) + geom_smooth(aes(color = ced.cat), se = FALSE, size = 1) +
facet_wrap(~sex)
## geom_smooth: method="auto" and size of largest group is >=1000, so using
## gam with formula: y ~ s(x, bs = "cs"). Use 'method = x' to change the
## smoothing method.
## geom_smooth: method="auto" and size of largest group is >=1000, so using
## gam with formula: y ~ s(x, bs = "cs"). Use 'method = x' to change the
## smoothing method.
Regression Models
- linear models:
lm()
- generalized linear models (probit, logit et al):
glm
- loads of packages on CRAN
- specify a model with a formula
mod.1 <- lm(formula = lndex ~ log(hhinc), data = dat)
summary(mod.1)
##
## Call:
## lm(formula = lndex ~ log(hhinc), data = dat)
##
## Residuals:
## Min 1Q Median 3Q Max
## -3.481 -0.307 -0.004 0.309 4.467
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 1.22476 0.05123 23.9 <2e-16 ***
## log(hhinc) 0.70251 0.00859 81.8 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.556 on 6776 degrees of freedom
## Multiple R-squared: 0.497, Adjusted R-squared: 0.497
## F-statistic: 6.7e+03 on 1 and 6776 DF, p-value: <2e-16
update() a model
- easy to iterate on models
mod.2 <- update(mod.1, . ~ . - 1) # leave formula as is, but substract the intercept
mod.3 <- update(mod.1, . ~ . + age + I(age^2)) # add age and age squared
coef(summary(mod.2)) # use coef() function to extract coefs only
## Estimate Std. Error t value Pr(>|t|)
## log(hhinc) 0.906 0.001179 768.5 0
coef(summary(mod.3))
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.9492539 7.710e-02 12.31 1.805e-34
## log(hhinc) 0.6422908 8.854e-03 72.55 0.000e+00
## age 0.0336750 2.667e-03 12.62 3.942e-36
## I(age^2) -0.0003756 2.525e-05 -14.88 2.767e-49
predict() data from a model
- use the
predict() function
- most regression functions have got their own prediction method.
- say we want to predict values of lndex from
mod.3
newdat <- expand.grid(hhinc = c(200, 300), age = c(20, 50, 80))
cbind(newdat, predict(mod.3, newdat))
## hhinc age predict(mod.3, newdat)
## 1 200 20 4.876
## 2 300 20 5.136
## 3 200 50 5.097
## 4 300 50 5.358
## 5 200 80 4.643
## 6 300 80 4.903
ggplot model coeficients
abline() is a convenient geom for this- works well for single explanatory variable
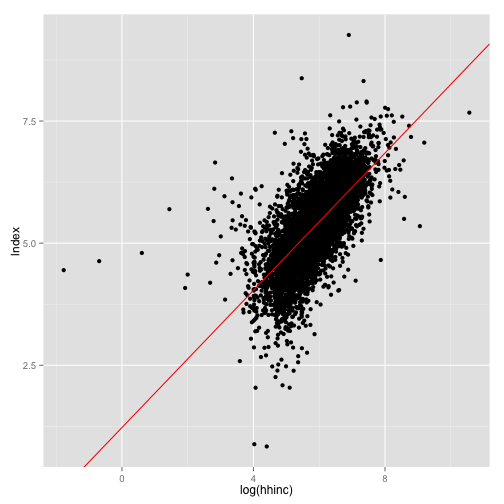
p <- ggplot(dat, aes(x = log(hhinc), y = lndex))
p + geom_point() + geom_abline(intercept = coef(mod.1)[1], slope = coef(mod.1)[2],
col = "red")
update() a model 2: add a dummy variable
- let’s add the number of rooms as a dummy variable
d
- i.e. we want d1=1 if nrooms==1, d1=0 else; d2=1 iff nrooms==2 etc
- R does that for you (you don’t want to create all those d’s)
mod.4 <- update(mod.3, . ~ . + factor(nrooms))
summary(mod.4)
##
## Call:
## lm(formula = lndex ~ log(hhinc) + age + I(age^2) + factor(nrooms),
## data = dat)
##
## Residuals:
## Min 1Q Median 3Q Max
## -3.751 -0.307 -0.008 0.303 3.477
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 9.09e-01 1.64e-01 5.56 2.9e-08 ***
## log(hhinc) 5.74e-01 9.36e-03 61.37 < 2e-16 ***
## age 2.66e-02 2.64e-03 10.09 < 2e-16 ***
## I(age^2) -3.16e-04 2.49e-05 -12.72 < 2e-16 ***
## factor(nrooms)2 3.37e-01 1.58e-01 2.13 0.03334 *
## factor(nrooms)3 3.12e-01 1.48e-01 2.11 0.03504 *
## factor(nrooms)4 5.27e-01 1.47e-01 3.59 0.00034 ***
## factor(nrooms)5 5.91e-01 1.47e-01 4.02 5.9e-05 ***
## factor(nrooms)6 7.53e-01 1.47e-01 5.12 3.1e-07 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.526 on 6769 degrees of freedom
## Multiple R-squared: 0.55, Adjusted R-squared: 0.549
## F-statistic: 1.03e+03 on 8 and 6769 DF, p-value: <2e-16
Hyothesis Testing, IV models
- check out
library(car) and library(AER)
- convenient functions for all applied econometrics tasks
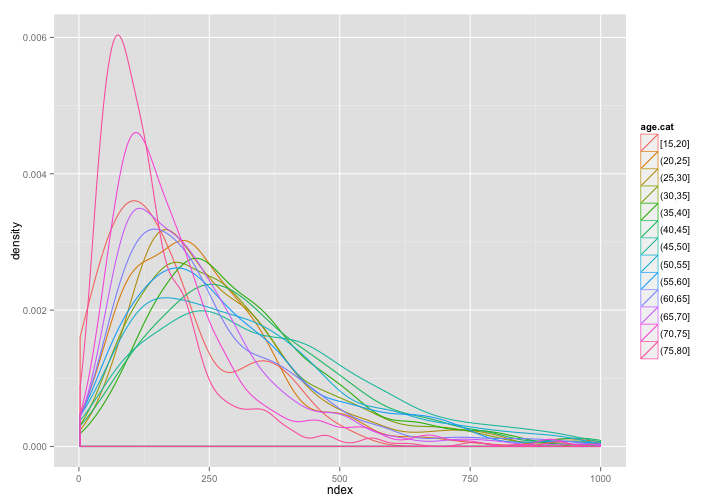
Create an Age Profile for consumption
- How does the distribution of non-durable expenditure vary wity age?
- How to model this in a regression model?
ggplot(subset(dat, ndex < 1000), aes(x = ndex)) + geom_density(aes(color = age.cat))
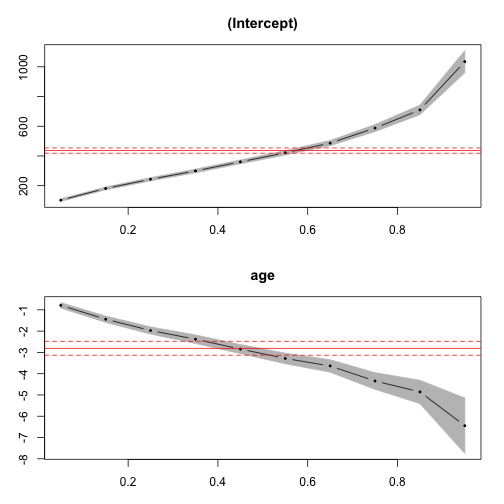
Age Profile: Quantile Regression
library(quantreg)
qreg50 <- rq(formula = ndex ~ age, data = dat, tau = 0.5) # median regression
qreg <- rq(formula = ndex ~ age, data = dat, tau = seq(0.05, 0.95,
le = 10)) # quantile reg at several quantiles
plot(summary(qreg)) # very nice plotting method
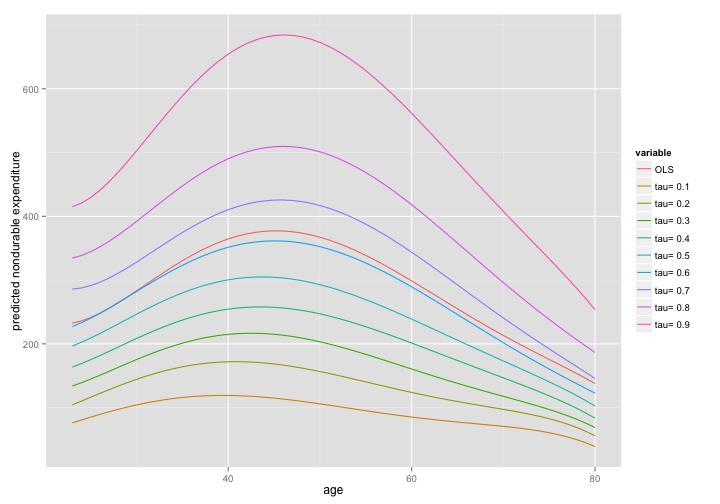
Age Profile: Compare OLS and Quantile Reg
- compare the Conditional Expectation (the OLS estimate) with the conditional quantile function
- fit a 5-th order polynomial in age
form <- ndex ~ age + I(age^2) + I(age^3) + I(age^4) + I(age^5)
lin.mod <- lm(formula = form, data=dat)
taus <- seq(0.1,0.9,le=9) # desired quantiles
qr.mod <- rq(formula= form,data=dat,tau=taus)
ages <- sort(unique(dat$age)) # vector of ages in data
# create data.frame with cols "age", and a prediction for each model
preds <- data.frame(age = ages, OLS= predict(lin.mod, newdata = data.frame(age = ages)))
# add qreg predictions
preds <- cbind(preds, predict(qr.mod,data.frame(age=ages)))
head(preds)
## age OLS tau= 0.1 tau= 0.2 tau= 0.3 tau= 0.4 tau= 0.5 tau= 0.6 tau= 0.7
## 1 17 267.1 51.39 78.01 122.5 150.5 186.3 221.6 336.6
## 2 18 252.6 54.85 80.65 121.0 149.3 183.9 217.6 318.8
## 3 19 242.2 58.70 84.21 121.2 149.7 183.4 215.9 305.4
## 4 20 235.4 62.85 88.51 122.8 151.5 184.7 216.2 295.8
## 5 21 231.7 67.20 93.42 125.5 154.5 187.4 218.4 289.6
## 6 22 230.9 71.66 98.77 129.3 158.6 191.5 222.1 286.5
## tau= 0.8 tau= 0.9
## 1 371.4 484.3
## 2 355.9 457.0
## 3 344.8 437.0
## 4 337.5 423.5
## 5 333.6 415.9
## 6 332.8 413.3
Plot OLS vs Quantile Reg Age Profiles
- ggplot works best with long data.
- need to
reshape preds into long
library(reshape)
mpreds <- melt(preds, id.vars = "age") # now the data is long
head(mpreds)
## age variable value
## 1 17 OLS 267.1
## 2 18 OLS 252.6
## 3 19 OLS 242.2
## 4 20 OLS 235.4
## 5 21 OLS 231.7
## 6 22 OLS 230.9
ggplot(subset(mpreds, age > 22), aes(x = age, y = value, color = variable)) +
geom_line() + scale_y_continuous(name = "predicted nondurable expenditure")
R and Time Series
- R has strong time series capabilities
- base time series architecture in
ts()
- more advanced ways to deal with time/date classes
- check out
data(UKDriverDeaths)
UK <- UKDriverDeaths
str(UK)
## Time-Series [1:192] from 1969 to 1985: 1687 1508 1507 1385 1632 ...
plot(UK)
plot(decompose(UK))
acf(UK)
pacf(UK)
R and Foreign Languages
- It is easy to call other languages from within R
- R is itself written in Fortran and C, so those two are particularly easy
- This is important for R users who want a lot of performance (MCMC, dynamic programming, etc)
R and Fortran
- suppose we have the fortran submodule
bar.f stored on disk
- it takes integer
n, makes x=1:n and returns x^2
- it looks like that
subroutine bar(n, x)
integer n
double precision x(n)
integer i
do 100 i = 1, n
x(i) = x(i) ** 2
100 continue
end
- we need to compile the function. write in your terminal
cd ~/git/R-demo
R CMD SHLIB bar.f
- It’s already compiled in the github repository so no need to if you downloaded that.
- finally, in R we need to load the dynamic library and call the function
dyn.load("bar.so")
how.many <- 5L # L means integer
y <- rnorm(how.many)
r.result <- y^2
# ideally we have a wrapper on .Fortran to check sanity of input
f.result <- .Fortran("bar", n=how.many, x=y) # .Fortran returns a list
identical(r.result,f.result$x) #identical?
## [1] TRUE
R and C/C++
library(Rcpp)- Rcpp Mastermind Dirk Eddelbuettel
- you can write packages the use C code via Rcpp
- very useful if you have a part of your code that needs speeding up.
- many derived libraries
library(RcppArmadillo): interface to the Armadillo C++ matrix algebra library. Armadillo is close to matlab.library(RcppEigen): interface to the C++ Eigen libraries. Eigen is a very versatile matrix algebra library. Great for sparse matrices.library(RcppSimpleTensor): High dimensional computations made easy- many more
- you can write C code inline and test it
R and C++ inline
library(inline) allows inline testing of C code
library(inline)
my.cpp.code <- "NumericVector xx(x);
return wrap( std::accumulate( xx.begin(), xx.end(), 0.0));"
my.cpp.fun <- cxxfunction( signature( x="numeric"), body=my.cpp.code, plugin="Rcpp")
y <- seq(1,10,0.5)
r.result <- sum(y)
c.result <- my.cpp.fun(y)
identical(r.result,c.result)
## [1] TRUE
# if you want to see what's going on under the hood:
my.cpp.fun <- cxxfunction( signature( x="numeric"), body=my.cpp.code, plugin="Rcpp", verbose=TRUE)
## >> setting environment variables:
## PKG_LIBS = /Library/Frameworks/R.framework/Versions/2.15/Resources/library/Rcpp/lib/x86_64/libRcpp.a
##
## >> LinkingTo : Rcpp
## CLINK_CPPFLAGS = -I"/Library/Frameworks/R.framework/Versions/2.15/Resources/library/Rcpp/include"
##
## >> Program source :
##
## 1 :
## 2 : // includes from the plugin
## 3 :
## 4 : #include <Rcpp.h>
## 5 :
## 6 :
## 7 : #ifndef BEGIN_RCPP
## 8 : #define BEGIN_RCPP
## 9 : #endif
## 10 :
## 11 : #ifndef END_RCPP
## 12 : #define END_RCPP
## 13 : #endif
## 14 :
## 15 : using namespace Rcpp;
## 16 :
## 17 :
## 18 : // user includes
## 19 :
## 20 :
## 21 : // declarations
## 22 : extern "C" {
## 23 : SEXP file90625393cbd( SEXP x) ;
## 24 : }
## 25 :
## 26 : // definition
## 27 :
## 28 : SEXP file90625393cbd( SEXP x ){
## 29 : BEGIN_RCPP
## 30 : NumericVector xx(x);
## 31 : return wrap( std::accumulate( xx.begin(), xx.end(), 0.0));
## 32 : END_RCPP
## 33 : }
## 34 :
## 35 :
## Compilation argument:
## /Library/Frameworks/R.framework/Resources/bin/R CMD SHLIB file90625393cbd.cpp 2> file90625393cbd.cpp.err.txt
How to make this set of slides
- Use RStudio to author an R markdown file
- click a button in RStudio to knit an HTML document
- this uses the amazing knitr package to “knit” (evaluated or not) R code into text documents.
- finally use pandoc to convert to HTML5 slides:
pandoc -s -S -t slidy --mathjax intro.md -o intro.html
- Get the R markdown source code of this presentation on github
- More info at MAGES blog