Software for download. All codes and software downloadable below are ©2008-2010
Paul Dalby.
Use of this software indicates that you agree not to copy or distribute it, or exploit it commercially without the authors permission.
PAD v7.0: (released 31 Aug 2010). Python based tool for guiding protein engineering and directed evolution strategies.
Download zip
file, unpack and follow Readme file to get started.
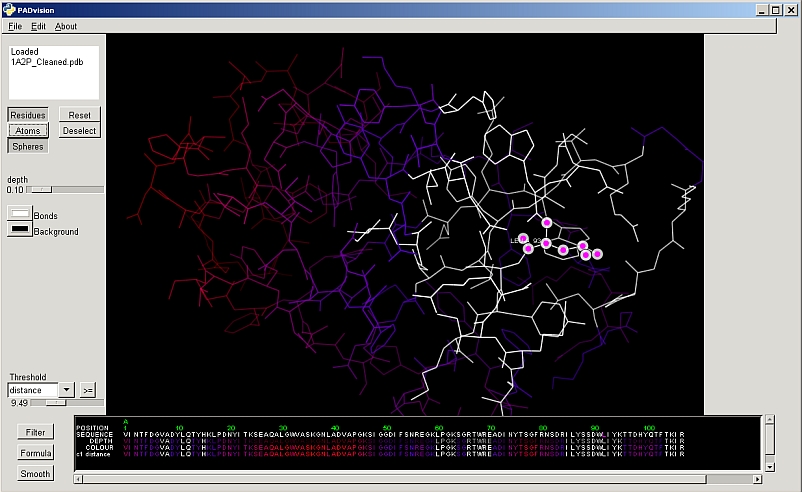
Calculates parameters for each residue and visualises by sequence and structure.
Creates formula-based calculations on parameters for each residue, or filter
desired parameters by boolean logic, then output to sequence and structure.
Parameters that can be calculated include:
- Distances to sets of residues/ligands
- Surface areas (all/non-polar), solvation energies
- Torsion energies
- Imported sequence entropies (eg. calculated from BioEdit)
- Protein and sequence Visualiser
- Ftp download of pdb files
- Bfactors

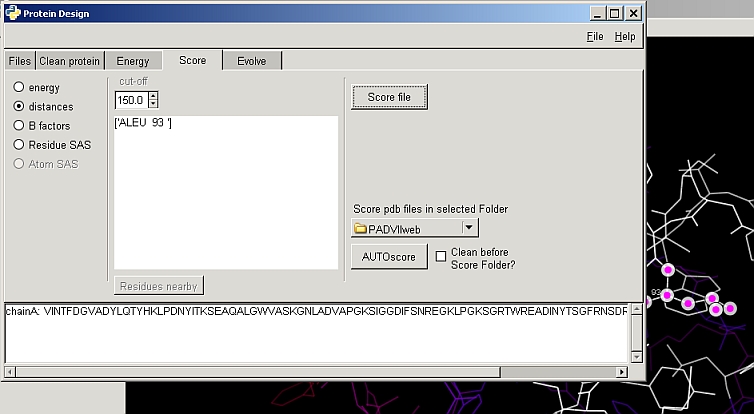
- Export all data to Excel
- Clean up and minimise missing residues
- Create pdb sphere of local residues (eg Theozymes)
- Create mutants and rotamer libraries
- Evaluate rotamer libraries and reject by steric clash
- Identify all steric clashes
Pdb files must be in the standard format.
download Buffer Calculator
Very simple buffer calculator. A much better one exists online here: http://www.liv.ac.uk/buffers/buffercalc.html